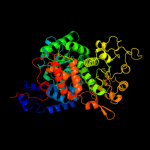
| 1 | c1o0sB_ |
|
 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent malic enzyme;
PDBTitle: crystal structure of ascaris suum malic enzyme complexed with nadh
|
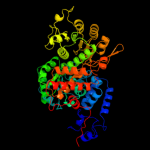
| 2 | c1gz3B_ |
|
 |
100.0 |
41 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent malic enzyme;
PDBTitle: molecular mechanism for the regulation of human mitochondrial2 nad(p)+-dependent malic enzyme by atp and fumarate
|
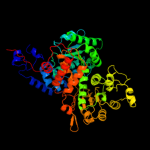
| 3 | c1qr6A_ |
|
 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malic enzyme 2;
PDBTitle: human mitochondrial nad(p)-dependent malic enzyme
|
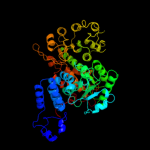
| 4 | c2aw5A_ |
|
 |
100.0 |
43 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent malic enzyme;
PDBTitle: crystal structure of a human malic enzyme
|
| 5 | c3nv9A_ |
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malic enzyme;
PDBTitle: crystal structure of entamoeba histolytica malic enzyme
|
| 6 | c1ww8A_ |
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate oxidoreductase;
PDBTitle: crystal structure of malic enzyme from pyrococcus2 horikoshii ot3
|
| 7 | c1vl6C_ |
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate oxidoreductase;
PDBTitle: crystal structure of nad-dependent malic enzyme (tm0542) from2 thermotoga maritima at 2.61 a resolution
|
| 8 | c2a9fB_ |
|
 |
100.0 |
30 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative malic enzyme ((s)-malate:nad+
PDBTitle: crystal structure of a putative malic enzyme ((s)-2 malate:nad+ oxidoreductase (decarboxylating))
|
| 9 | d1o0sa2 |
|
 |
100.0 |
45 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Malic enzyme N-domain |
| 10 | d1pj3a2 |
|
 |
100.0 |
46 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Malic enzyme N-domain |
| 11 | d1gq2a2 |
|
 |
100.0 |
48 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Malic enzyme N-domain |
| 12 | d1o0sa1 |
|
 |
100.0 |
35 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 13 | d1pj3a1 |
|
 |
100.0 |
34 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 14 | d1gq2a1 |
|
 |
100.0 |
36 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 15 | d1vl6a1 |
|
 |
100.0 |
31 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 16 | d1vl6a2 |
|
 |
100.0 |
24 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Malic enzyme N-domain |
| 17 | d1euza1 |
|
 |
98.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 18 | d1li4a1 |
|
 |
98.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 19 | c3n58D_ |
|
 |
97.7 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
| 20 | d1v9la1 |
|
 |
97.6 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 21 | d1v8ba1 |
|
not modelled |
97.4 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 22 | c3aogA_ |
|
not modelled |
97.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase;
PDBTitle: crystal structure of glutamate dehydrogenase (gdhb) from thermus2 thermophilus (glu bound form)
|
| 23 | c3dhyC_ |
|
not modelled |
97.3 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
|
| 24 | d1uxja1 |
|
not modelled |
97.2 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 25 | c3gvpB_ |
|
not modelled |
97.2 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenosylhomocysteinase 3;
PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
|
| 26 | c2rirA_ |
|
not modelled |
97.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 27 | c2bmaA_ |
|
not modelled |
97.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase (nadp+);
PDBTitle: the crystal structure of plasmodium falciparum glutamate2 dehydrogenase, a putative target for novel antimalarial3 drugs
|
| 28 | c3d4oA_ |
|
not modelled |
97.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 29 | c1d4fD_ |
|
not modelled |
97.0 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:s-adenosylhomocysteine hydrolase;
PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
|
| 30 | d1t2da1 |
|
not modelled |
97.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 31 | c3aoeC_ |
|
not modelled |
96.9 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glutamate dehydrogenase;
PDBTitle: crystal structure of hetero-hexameric glutamate dehydrogenase from2 thermus thermophilus (leu bound form)
|
| 32 | d1pzga1 |
|
not modelled |
96.8 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 33 | d1gpja2 |
|
not modelled |
96.7 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 34 | d1l7da1 |
|
not modelled |
96.7 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 35 | c1v9lA_ |
|
not modelled |
96.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase;
PDBTitle: l-glutamate dehydrogenase from pyrobaculum islandicum2 complexed with nad
|
| 36 | d1u8xx1 |
|
not modelled |
96.6 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 37 | c1pzfD_ |
|
not modelled |
96.6 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:lactate dehydrogenase;
PDBTitle: t.gondii ldh1 ternary complex with apad+ and oxalate
|
| 38 | c3d64A_ |
|
not modelled |
96.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
| 39 | d1bvua1 |
|
not modelled |
96.5 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 40 | c1gpjA_ |
|
not modelled |
96.5 |
21 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
| 41 | d1vi2a1 |
|
not modelled |
96.5 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 42 | c1bvuF_ |
|
not modelled |
96.4 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:protein (glutamate dehydrogenase);
PDBTitle: glutamate dehydrogenase from thermococcus litoralis
|
| 43 | c1obbB_ |
|
not modelled |
96.4 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-glucosidase;
PDBTitle: alpha-glucosidase a, agla, from thermotoga maritima in2 complex with maltose and nad+
|
| 44 | c1nr1A_ |
|
not modelled |
96.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase 1;
PDBTitle: crystal structure of the r463a mutant of human glutamate2 dehydrogenase
|
| 45 | c1pjcA_ |
|
not modelled |
96.2 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (l-alanine dehydrogenase);
PDBTitle: l-alanine dehydrogenase complexed with nad
|
| 46 | d1s6ya1 |
|
not modelled |
96.2 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 47 | c3tozA_ |
|
not modelled |
96.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
|
| 48 | d1llda1 |
|
not modelled |
96.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 49 | c1hrdA_ |
|
not modelled |
96.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase;
PDBTitle: glutamate dehydrogenase
|
| 50 | c2fnzA_ |
|
not modelled |
96.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of the lactate dehydrogenase from cryptosporidium2 parvum complexed with cofactor (b-nicotinamide adenine dinucleotide)3 and inhibitor (oxamic acid)
|
| 51 | c3fefB_ |
|
not modelled |
96.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative glucosidase lpld;
PDBTitle: crystal structure of putative glucosidase lpld from2 bacillus subtilis
|
| 52 | c2eezG_ |
|
not modelled |
96.0 |
17 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase from themus thermophilus
|
| 53 | d1a4ia1 |
|
not modelled |
95.8 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 54 | d1i10a1 |
|
not modelled |
95.8 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 55 | c3p2yA_ |
|
not modelled |
95.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alanine dehydrogenase/pyridine nucleotide transhydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
|
| 56 | c1u8xX_ |
|
not modelled |
95.7 |
17 |
PDB header:hydrolase
Chain: X: PDB Molecule:maltose-6'-phosphate glucosidase;
PDBTitle: crystal structure of glva from bacillus subtilis, a metal-requiring,2 nad-dependent 6-phospho-alpha-glucosidase
|
| 57 | d9ldta1 |
|
not modelled |
95.6 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 58 | d1obba1 |
|
not modelled |
95.6 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 59 | d1vjta1 |
|
not modelled |
95.6 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 60 | c2ep9A_ |
|
not modelled |
95.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-gulonate 3-dehydrogenase;
PDBTitle: crystal structure of the rabbit l-gulonate 3-dehydrogenase2 (nadh form)
|
| 61 | d1b0aa1 |
|
not modelled |
95.6 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 62 | d1ldna1 |
|
not modelled |
95.5 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 63 | c1v8bA_ |
|
not modelled |
95.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of a hydrolase
|
| 64 | c3l07B_ |
|
not modelled |
95.4 |
22 |
PDB header:oxidoreductase,hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
|
| 65 | d1gtma1 |
|
not modelled |
95.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 66 | d1ojua1 |
|
not modelled |
95.4 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 67 | c1a4iB_ |
|
not modelled |
95.4 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
| 68 | d1guza1 |
|
not modelled |
95.3 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 69 | c1ur5C_ |
|
not modelled |
95.3 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: stabilization of a tetrameric malate dehydrogenase by2 introduction of a disulfide bridge at the dimer/dimer3 interface
|
| 70 | c2hjrK_ |
|
not modelled |
95.3 |
21 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
| 71 | d1pjca1 |
|
not modelled |
95.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 72 | d1ez4a1 |
|
not modelled |
95.3 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 73 | c3oneA_ |
|
not modelled |
95.2 |
20 |
PDB header:hydrolase/hydrolase substrate
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
|
| 74 | d1b26a1 |
|
not modelled |
95.2 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 75 | c1u4sA_ |
|
not modelled |
95.2 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: plasmodium falciparum lactate dehydrogenase complexed with 2,6-2 naphthalenedisulphonic acid
|
| 76 | c1wwkA_ |
|
not modelled |
95.1 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
| 77 | d1a5za1 |
|
not modelled |
95.1 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 78 | c2tmgD_ |
|
not modelled |
95.1 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:protein (glutamate dehydrogenase);
PDBTitle: thermotoga maritima glutamate dehydrogenase mutant s128r,2 t158e, n117r, s160e
|
| 79 | c2v6bB_ |
|
not modelled |
95.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of lactate dehydrogenase from deinococcus2 radiodurans (apo form)
|
| 80 | d1gv0a1 |
|
not modelled |
95.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 81 | d1y6ja1 |
|
not modelled |
95.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 82 | c2j6iC_ |
|
not modelled |
95.0 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:formate dehydrogenase;
PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal2 mutant
|
| 83 | c3evtA_ |
|
not modelled |
95.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from2 lactobacillus plantarum
|
| 84 | c4a5oB_ |
|
not modelled |
95.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of pseudomonas aeruginosa n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
| 85 | c1s6yA_ |
|
not modelled |
94.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phospho-beta-glucosidase;
PDBTitle: 2.3a crystal structure of phospho-beta-glucosidase
|
| 86 | c3d0oA_ |
|
not modelled |
94.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase 1;
PDBTitle: crystal structure of lactate dehydrogenase from2 staphylococcus aureus
|
| 87 | c3p2oA_ |
|
not modelled |
94.7 |
24 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 88 | c3p2oB_ |
|
not modelled |
94.5 |
23 |
PDB header:oxidoreductase, hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 89 | c4a26B_ |
|
not modelled |
94.5 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative c-1-tetrahydrofolate synthase, cytoplasmic;
PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
|
| 90 | c2cukC_ |
|
not modelled |
94.5 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
| 91 | c1ez4B_ |
|
not modelled |
94.5 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of non-allosteric l-lactate dehydrogenase2 from lactobacillus pentosus at 2.3 angstrom resolution
|
| 92 | c3gviB_ |
|
not modelled |
94.5 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of lactate/malate dehydrogenase from2 brucella melitensis in complex with adp
|
| 93 | c3nepX_ |
|
not modelled |
94.4 |
24 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:malate dehydrogenase;
PDBTitle: 1.55a resolution structure of malate dehydrogenase from salinibacter2 ruber
|
| 94 | c3k8zD_ |
|
not modelled |
94.4 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nad-specific glutamate dehydrogenase;
PDBTitle: crystal structure of gudb1 a decryptified secondary glutamate2 dehydrogenase from b. subtilis
|
| 95 | d1hwxa1 |
|
not modelled |
94.4 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 96 | d1llca1 |
|
not modelled |
94.3 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 97 | d1i0za1 |
|
not modelled |
94.3 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 98 | d1hyha1 |
|
not modelled |
94.2 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 99 | c3donA_ |
|
not modelled |
94.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from staphylococcus2 epidermidis
|
| 100 | d1bgva1 |
|
not modelled |
94.0 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 101 | d1jw9b_ |
|
not modelled |
94.0 |
16 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Molybdenum cofactor biosynthesis protein MoeB |
| 102 | c3u62A_ |
|
not modelled |
93.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from thermotoga maritima
|
| 103 | c2gcgB_ |
|
not modelled |
93.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
| 104 | c1zfnA_ |
|
not modelled |
93.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:adenylyltransferase thif;
PDBTitle: structural analysis of escherichia coli thif
|
| 105 | c2hk8B_ |
|
not modelled |
93.9 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
|
| 106 | d5ldha1 |
|
not modelled |
93.8 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 107 | c2d4aC_ |
|
not modelled |
93.6 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of the malate dehydrogenase from aeropyrum pernix
|
| 108 | c1up6F_ |
|
not modelled |
93.6 |
25 |
PDB header:hydrolase
Chain: F: PDB Molecule:6-phospho-beta-glucosidase;
PDBTitle: structure of the 6-phospho-beta glucosidase from thermotoga2 maritima at 2.55 angstrom resolution in the tetragonal form3 with manganese, nad+ and glucose-6-phosphate
|
| 109 | c1vjtA_ |
|
not modelled |
93.6 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-glucosidase;
PDBTitle: crystal structure of alpha-glucosidase (tm0752) from thermotoga2 maritima at 2.50 a resolution
|
| 110 | c1ojuA_ |
|
not modelled |
93.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: 2.8 a resolution structure of malate dehydrogenase from2 archaeoglobus fulgidus in complex with etheno-nad.
|
| 111 | c1b0aA_ |
|
not modelled |
93.4 |
22 |
PDB header:oxidoreductase,hydrolase
Chain: A: PDB Molecule:protein (fold bifunctional protein);
PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
|
| 112 | c3h9gA_ |
|
not modelled |
93.4 |
14 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:mccb protein;
PDBTitle: crystal structure of e. coli mccb + mcca-n7isoasn
|
| 113 | c3gucB_ |
|
not modelled |
93.3 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:ubiquitin-like modifier-activating enzyme 5;
PDBTitle: human ubiquitin-activating enzyme 5 in complex with amppnp
|
| 114 | c8ldhA_ |
|
not modelled |
93.3 |
24 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:m4 apo-lactate dehydrogenase;
PDBTitle: refined crystal structure of dogfish m4 apo-lactate2 dehydrogenase
|
| 115 | c1luaA_ |
|
not modelled |
93.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methylene tetrahydromethanopterin dehydrogenase;
PDBTitle: structure of methylene-tetrahydromethanopterin dehydrogenase from2 methylobacterium extorquens am1 complexed with nadp
|
| 116 | c1ldbA_ |
|
not modelled |
93.2 |
17 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:apo-l-lactate dehydrogenase;
PDBTitle: structure determination and refinement of bacillus2 stearothermophilus lactate dehydrogenase
|
| 117 | d1ldma1 |
|
not modelled |
93.1 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 118 | d1luaa1 |
|
not modelled |
93.1 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 119 | d1mlda1 |
|
not modelled |
93.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 120 | c3vh1A_ |
|
not modelled |
93.0 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: crystal structure of saccharomyces cerevisiae atg7 (1-595)
|

