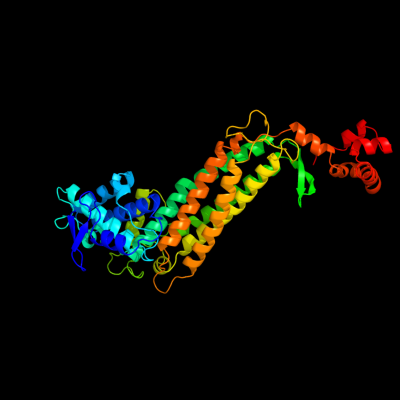
| 1 | d1j3ua_
|
|
 |
100.0 |
41 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
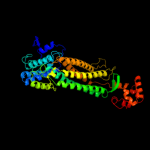
| 2 | d1vdka_
|
|
 |
100.0 |
58 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
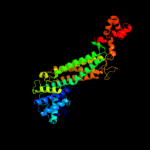
| 3 | c1yfmA_
|
|
 |
100.0 |
59 |
PDB header:lyase
Chain: A: PDB Molecule:fumarase;
PDBTitle: recombinant yeast fumarase
|
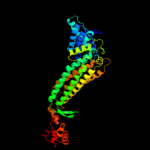
| 4 | d1yfma_
|
|
 |
100.0 |
59 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 5 | d1fuoa_
|
|
 |
100.0 |
100 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 6 | d1jswa_
|
|
 |
100.0 |
40 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 7 | c3no9C_
|
|
 |
100.0 |
48 |
PDB header:lyase
Chain: C: PDB Molecule:fumarate hydratase class ii;
PDBTitle: crystal structure of apo fumarate hydratase from mycobacterium2 tuberculosis
|
| 8 | c3ocfB_
|
|
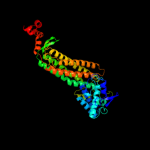 |
100.0 |
45 |
PDB header:lyase
Chain: B: PDB Molecule:fumarate lyase:delta crystallin;
PDBTitle: crystal structure of fumarate lyase:delta crystallin from brucella2 melitensis in native form
|
| 9 | c3e04C_
|
|
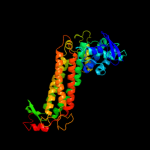 |
100.0 |
59 |
PDB header:lyase
Chain: C: PDB Molecule:fumarate hydratase;
PDBTitle: crystal structure of human fumarate hydratase
|
| 10 | c3gtdB_
|
|
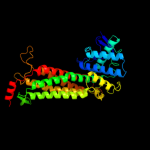 |
100.0 |
56 |
PDB header:lyase
Chain: B: PDB Molecule:fumarate hydratase class ii;
PDBTitle: 2.4 angstrom crystal structure of fumarate hydratase from rickettsia2 prowazekii
|
| 11 | d1jswc_
|
|
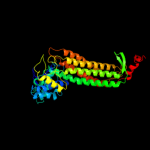 |
100.0 |
41 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 12 | c3r6yG_
|
|
 |
100.0 |
41 |
PDB header:lyase
Chain: G: PDB Molecule:aspartase;
PDBTitle: crystal structure of chymotrypsin-treated aspartase from bacillus sp.2 ym55-1
|
| 13 | c2pfmA_
|
|
 |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:adenylosuccinate lyase;
PDBTitle: crystal structure of adenylosuccinate lyase (purb) from bacillus2 anthracis
|
| 14 | d1tj7a_
|
|
 |
100.0 |
20 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 15 | d1re5a_
|
|
 |
100.0 |
24 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 16 | d1q5na_
|
|
 |
100.0 |
22 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 17 | c2e9fC_
|
|
 |
100.0 |
24 |
PDB header:lyase
Chain: C: PDB Molecule:argininosuccinate lyase;
PDBTitle: crystal structure of t.th.hb8 argininosuccinate lyase complexed with2 l-arginine
|
| 18 | d1c3ca_
|
|
 |
100.0 |
20 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 19 | d1i0aa_
|
|
 |
100.0 |
21 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 20 | d1tjva_
|
|
 |
100.0 |
21 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 21 | d1hy0a_ |
|
not modelled |
100.0 |
20 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 22 | d1k62a_ |
|
not modelled |
100.0 |
20 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 23 | c3c8tA_ |
|
not modelled |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:fumarate lyase;
PDBTitle: crystal structure of fumarate lyase from mesorhizobium sp. bnc1
|
| 24 | c3bhgA_ |
|
not modelled |
100.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:adenylosuccinate lyase;
PDBTitle: crystal structure of adenylosuccinate lyase from legionella2 pneumophila
|
| 25 | c2vd6B_ |
|
not modelled |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:adenylosuccinate lyase;
PDBTitle: human adenylosuccinate lyase in complex with its substrate2 n6-(1,2-dicarboxyethyl)-amp, and its products amp and3 fumarate.
|
| 26 | c2qgaC_ |
|
not modelled |
100.0 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:adenylosuccinate lyase;
PDBTitle: plasmodium vivax adenylosuccinate lyase pv003765 with amp bound
|
| 27 | c2ptsA_ |
|
not modelled |
100.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:adenylosuccinate lyase;
PDBTitle: crystal structure of wild type escherichia coli adenylosuccinate lyase
|
| 28 | d1dofa_ |
|
not modelled |
100.0 |
21 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 29 | c1yisA_ |
|
not modelled |
100.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:adenylosuccinate lyase;
PDBTitle: structural genomics of caenorhabditis elegans: adenylosuccinate lyase
|
| 30 | c2fenA_ |
|
not modelled |
100.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:3-carboxy-cis,cis-muconate lactonizing enzyme;
PDBTitle: 3-carboxy-cis,cis-muconate lactonizing enzyme from agrobacterium2 radiobacter s2
|
| 31 | d1f1oa_ |
|
not modelled |
100.0 |
19 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 32 | d1gkma_ |
|
not modelled |
52.1 |
19 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:HAL/PAL-like |
| 33 | c3exmA_ |
|
not modelled |
27.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphatase sc4828;
PDBTitle: crystal structure of the phosphatase sc4828 with the non-hydrolyzable2 nucleotide gpcp
|
| 34 | c2xgvA_ |
|
not modelled |
25.7 |
27 |
PDB header:viral protein
Chain: A: PDB Molecule:psiv capsid n-terminal domain;
PDBTitle: structure of the n-terminal domain of capsid protein from2 rabbit endogenous lentivirus (relik)
|
| 35 | d1lbua1 |
|
not modelled |
25.1 |
21 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:Peptidoglycan binding domain, PGBD |
| 36 | d1h8ba_ |
|
not modelled |
22.2 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:EF-hand modules in multidomain proteins |
| 37 | d2ce7a1 |
|
not modelled |
20.5 |
21 |
Fold:FtsH protease domain-like
Superfamily:FtsH protease domain-like
Family:FtsH protease domain-like |
| 38 | d1e8ga1 |
|
not modelled |
19.8 |
21 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
| 39 | c2ctoA_ |
|
not modelled |
19.8 |
42 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:novel protein;
PDBTitle: solution structure of the hmg box like domain from human2 hypothetical protein flj14904
|
| 40 | d1wvfa1 |
|
not modelled |
19.3 |
16 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
| 41 | c2di4B_ |
|
not modelled |
19.1 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:cell division protein ftsh homolog;
PDBTitle: crystal structure of the ftsh protease domain
|
| 42 | d2di4a1 |
|
not modelled |
18.5 |
16 |
Fold:FtsH protease domain-like
Superfamily:FtsH protease domain-like
Family:FtsH protease domain-like |
| 43 | c3bkhA_ |
|
not modelled |
16.2 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:lytic transglycosylase;
PDBTitle: crystal structure of the bacteriophage phikz lytic2 transglycosylase, gp144
|
| 44 | c3lq9B_ |
|
not modelled |
15.7 |
21 |
PDB header:signaling protein
Chain: B: PDB Molecule:dna-damage-inducible transcript 4 protein;
PDBTitle: crystal strucure of human redd1, a hypoxia-induced regulator2 of mtor
|
| 45 | c3bbnC_ |
|
not modelled |
14.3 |
40 |
PDB header:ribosome
Chain: C: PDB Molecule:ribosomal protein s3;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 46 | c2zttA_ |
|
not modelled |
12.8 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:rna-directed rna polymerase catalytic subunit;
PDBTitle: crystal structure of rna polymerase pb1-pb2 subunits from2 influenza a virus
|
| 47 | d3e9va1 |
|
not modelled |
12.7 |
22 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 48 | c3fbtB_ |
|
not modelled |
11.6 |
20 |
PDB header:oxidoreductase, lyase
Chain: B: PDB Molecule:chorismate mutase and shikimate 5-dehydrogenase
PDBTitle: crystal structure of a chorismate mutase/shikimate 5-2 dehydrogenase fusion protein from clostridium3 acetobutylicum
|
| 49 | c2ev9B_ |
|
not modelled |
11.5 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from thermus2 thermophilus hb8 in complex with nadp(h) and shikimate
|
| 50 | c1byvA_ |
|
not modelled |
10.7 |
30 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:protein (calcitonin);
PDBTitle: glycosylated eel calcitonin
|
| 51 | d1nvta2 |
|
not modelled |
10.7 |
13 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 52 | d1npya2 |
|
not modelled |
10.5 |
50 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 53 | d1n7ka_ |
|
not modelled |
9.8 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 54 | c1htrP_ |
|
not modelled |
9.2 |
14 |
PDB header:aspartyl protease
Chain: P: PDB Molecule:progastricsin (pro segment);
PDBTitle: crystal and molecular structures of human progastricsin at 1.622 angstroms resolution
|
| 55 | d1vi2a2 |
|
not modelled |
9.2 |
14 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 56 | c1lbuA_ |
|
not modelled |
8.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:muramoyl-pentapeptide carboxypeptidase;
PDBTitle: hydrolase metallo (zn) dd-peptidase
|
| 57 | d2z15a1 |
|
not modelled |
8.4 |
13 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 58 | c1wjvA_ |
|
not modelled |
8.4 |
44 |
PDB header:dna binding protein
Chain: A: PDB Molecule:cell growth regulating nucleolar protein lyar;
PDBTitle: solution structure of the n-terminal zinc finger domain of2 mouse cell growth regulating nucleolar protein lyar
|
| 59 | d1wjfa_ |
|
not modelled |
8.3 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:N-terminal Zn binding domain of HIV integrase
Family:N-terminal Zn binding domain of HIV integrase |
| 60 | c3m1eA_ |
|
not modelled |
8.2 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: crystal structure of benm_dbd
|
| 61 | d1k6ya1 |
|
not modelled |
8.1 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:N-terminal Zn binding domain of HIV integrase
Family:N-terminal Zn binding domain of HIV integrase |
| 62 | c1nvtA_ |
|
not modelled |
8.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5'-dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe or2 mj1084) in complex with nadp+
|
| 63 | c3no7A_ |
|
not modelled |
7.9 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative plasmid related protein;
PDBTitle: crystal structure of the centromere-binding protein parb from plasmid2 pcxc100
|
| 64 | d1e0ea_ |
|
not modelled |
7.8 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:N-terminal Zn binding domain of HIV integrase
Family:N-terminal Zn binding domain of HIV integrase |
| 65 | c1yrlD_ |
|
not modelled |
7.8 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: escherichia coli ketol-acid reductoisomerase
|
| 66 | d1rxqa_ |
|
not modelled |
7.5 |
16 |
Fold:DinB/YfiT-like putative metalloenzymes
Superfamily:DinB/YfiT-like putative metalloenzymes
Family:YfiT-like putative metal-dependent hydrolases |
| 67 | d2qalc1 |
|
not modelled |
7.1 |
38 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Prokaryotic type KH domain (KH-domain type II)
Family:Prokaryotic type KH domain (KH-domain type II) |
| 68 | d1p77a2 |
|
not modelled |
7.1 |
14 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 69 | c3pwzA_ |
|
not modelled |
6.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase 3;
PDBTitle: crystal structure of an ael1 enzyme from pseudomonas putida
|
| 70 | d2b0ja1 |
|
not modelled |
6.8 |
21 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:HMD dimerization domain-like |
| 71 | c2dxbR_ |
|
not modelled |
6.7 |
15 |
PDB header:hydrolase
Chain: R: PDB Molecule:thiocyanate hydrolase subunit gamma;
PDBTitle: recombinant thiocyanate hydrolase comprising partially-modified cobalt2 centers
|
| 72 | c3u62A_ |
|
not modelled |
6.7 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from thermotoga maritima
|
| 73 | d2d28c1 |
|
not modelled |
6.6 |
28 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:EspE N-terminal domain-like
Family:GSPII protein E N-terminal domain-like |
| 74 | d1nyta2 |
|
not modelled |
6.6 |
14 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 75 | c3pmoA_ |
|
not modelled |
6.6 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-acyltransferase;
PDBTitle: the structure of lpxd from pseudomonas aeruginosa at 1.3 a resolution
|
| 76 | c2nloA_ |
|
not modelled |
6.4 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of the quinate dehydrogenase from corynebacterium2 glutamicum
|
| 77 | c1p74B_ |
|
not modelled |
6.4 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe) from2 haemophilus influenzae
|
| 78 | d1u5ta1 |
|
not modelled |
6.3 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Vacuolar sorting protein domain |
| 79 | c2w6aB_ |
|
not modelled |
6.3 |
17 |
PDB header:signaling protein
Chain: B: PDB Molecule:arf gtpase-activating protein git1;
PDBTitle: x-ray structure of the dimeric git1 coiled-coil domain
|
| 80 | c3tozA_ |
|
not modelled |
6.3 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
|
| 81 | c2eggA_ |
|
not modelled |
6.3 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from2 geobacillus kaustophilus
|
| 82 | d2dy1a3 |
|
not modelled |
6.3 |
22 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 83 | c1ahuB_ |
|
not modelled |
6.1 |
21 |
PDB header:flavoenzyme
Chain: B: PDB Molecule:vanillyl-alcohol oxidase;
PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
|
| 84 | c2dinA_ |
|
not modelled |
6.1 |
40 |
PDB header:dna binding protein
Chain: A: PDB Molecule:cell division cycle 5-like protein;
PDBTitle: solution structure of the myb_dna-binding domain of human2 cell division cycle 5-like protein
|
| 85 | d2uubc1 |
|
not modelled |
6.1 |
33 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Prokaryotic type KH domain (KH-domain type II)
Family:Prokaryotic type KH domain (KH-domain type II) |
| 86 | c2hk8B_ |
|
not modelled |
6.0 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
|
| 87 | d1ugpa_ |
|
not modelled |
6.0 |
38 |
Fold:Nitrile hydratase alpha chain
Superfamily:Nitrile hydratase alpha chain
Family:Nitrile hydratase alpha chain |
| 88 | d1g3pa2 |
|
not modelled |
6.0 |
20 |
Fold:N-terminal domains of the minor coat protein g3p
Superfamily:N-terminal domains of the minor coat protein g3p
Family:N-terminal domains of the minor coat protein g3p |
| 89 | c3pgjB_ |
|
not modelled |
5.9 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.49 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae o1 biovar eltor str. n169613 in complex with shikimate
|
| 90 | d2nr7a1 |
|
not modelled |
5.9 |
23 |
Fold:Lysozyme-like
Superfamily:Lysozyme-like
Family:NMB1012-like |
| 91 | c2hinA_ |
|
not modelled |
5.8 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:repressor protein;
PDBTitle: structure of n15 cro at 1.05 a: an ortholog of lambda cro2 with a completely different but equally effective3 dimerization mechanism
|
| 92 | d2bv3a3 |
|
not modelled |
5.7 |
25 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 93 | c3iymA_ |
|
not modelled |
5.7 |
13 |
PDB header:virus
Chain: A: PDB Molecule:capsid protein;
PDBTitle: backbone trace of the capsid protein dimer of a fungal partitivirus2 from electron cryomicroscopy and homology modeling
|
| 94 | c2fgyA_ |
|
not modelled |
5.6 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:carboxysome shell polypeptide;
PDBTitle: beta carbonic anhydrase from the carboxysomal shell of2 halothiobacillus neapolitanus (csosca)
|
| 95 | c2zmeA_ |
|
not modelled |
5.6 |
17 |
PDB header:protein transport
Chain: A: PDB Molecule:vacuolar-sorting protein snf8;
PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
|
| 96 | c1x5bA_ |
|
not modelled |
5.5 |
56 |
PDB header:protein binding
Chain: A: PDB Molecule:signal transducing adaptor molecule 2;
PDBTitle: the solution structure of the vhs domain of human signal2 transducing adaptor molecule 2
|
| 97 | c1npyA_ |
|
not modelled |
5.5 |
43 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical shikimate 5-dehydrogenase-like
PDBTitle: structure of shikimate 5-dehydrogenase-like protein hi0607
|
| 98 | c2kheA_ |
|
not modelled |
5.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:toxin-like protein;
PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
|
| 99 | c3hzsA_ |
|
not modelled |
5.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:monofunctional glycosyltransferase;
PDBTitle: s. aureus monofunctional glycosyltransferase (mtga)in complex with2 moenomycin
|