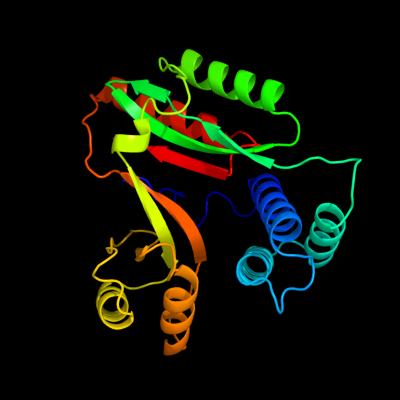
| 1 | d1lfpa_
|
|
 |
100.0 |
36 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
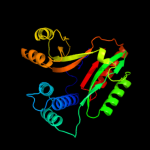
| 2 | d1kona_
|
|
 |
100.0 |
39 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
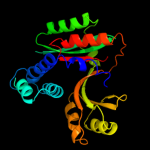
| 3 | d1mw7a_
|
|
 |
100.0 |
32 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
| 4 | c2jsxA_
|
|
 |
58.9 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:protein napd;
PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
|
| 5 | c2zf8A_
|
|
 |
43.0 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:component of sodium-driven polar flagellar motor;
PDBTitle: crystal structure of moty
|
| 6 | c2vsaA_
|
|
 |
35.3 |
55 |
PDB header:toxin
Chain: A: PDB Molecule:mosquitocidal toxin;
PDBTitle: structure and mode of action of a mosquitocidal holotoxin
|
| 7 | c2dhaA_
|
|
 |
35.2 |
12 |
PDB header:rna binding protein
Chain: A: PDB Molecule:flj20171 protein;
PDBTitle: solution structure of the second rna recognition motif in2 hypothetical protein flj201171
|
| 8 | d2cyya2
|
|
 |
33.9 |
10 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 9 | c3ibwA_
|
|
 |
32.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:gtp pyrophosphokinase;
PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
|
| 10 | c2zbcH_
|
|
 |
32.6 |
12 |
PDB header:transcription
Chain: H: PDB Molecule:83aa long hypothetical transcriptional regulator asnc;
PDBTitle: crystal structure of sts042, a stand-alone ram module protein, from2 hyperthermophilic archaeon sulfolobus tokodaii strain7.
|
| 11 | d1kkha2
|
|
 |
31.9 |
14 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Mevalonate kinase |
| 12 | c1fpqA_
|
|
 |
31.6 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:isoliquiritigenin 2'-o-methyltransferase;
PDBTitle: crystal structure analysis of selenomethionine substituted chalcone o-2 methyltransferase
|
| 13 | d1ewqa2
|
|
 |
31.4 |
39 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 14 | d1weya_
|
|
 |
31.3 |
17 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 15 | c2nyiB_
|
|
 |
31.2 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:unknown protein;
PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
|
| 16 | d1p65a_
|
|
 |
31.0 |
26 |
Fold:Nucleocapsid protein dimerization domain
Superfamily:Nucleocapsid protein dimerization domain
Family:Arterivirus nucleocapsid protein |
| 17 | c1p65A_
|
|
 |
31.0 |
26 |
PDB header:viral protein
Chain: A: PDB Molecule:nucleocapsid protein;
PDBTitle: crystal structure of the nucleocapsid protein of porcine reproductive2 and respiratory syndrome virus (prrsv)
|
| 18 | c3k2qA_
|
|
 |
29.2 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:pyrophosphate-dependent phosphofructokinase;
PDBTitle: crystal structure of pyrophosphate-dependent2 phosphofructokinase from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr88
|
| 19 | d2f1fa2
|
|
 |
26.5 |
14 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 20 | d2pc6a1
|
|
 |
25.4 |
20 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 21 | d2ex4a1 |
|
not modelled |
23.4 |
30 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:AD-003 protein-like |
| 22 | d1ub9a_ |
|
not modelled |
23.1 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 23 | d1k47a2 |
|
not modelled |
22.9 |
10 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Phosphomevalonate kinase (PMK) |
| 24 | c2b8kD_ |
|
not modelled |
22.9 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:dna-directed rna polymerase ii 32 kda
PDBTitle: 12-subunit rna polymerase ii
|
| 25 | c3l7pA_ |
|
not modelled |
22.3 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:putative nitrogen regulatory protein pii;
PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
|
| 26 | c3s01A_ |
|
not modelled |
22.2 |
14 |
PDB header:rna binding protein
Chain: A: PDB Molecule:heterogeneous nuclear ribonucleoprotein l;
PDBTitle: crystal structure of a heterogeneous nuclear ribonucleoprotein l2 (hnrpl) from mus musculus at 2.15 a resolution
|
| 27 | d1kkca2 |
|
not modelled |
22.2 |
32 |
Fold:Fe,Mn superoxide dismutase (SOD), C-terminal domain
Superfamily:Fe,Mn superoxide dismutase (SOD), C-terminal domain
Family:Fe,Mn superoxide dismutase (SOD), C-terminal domain |
| 28 | d1gtra1 |
|
not modelled |
22.0 |
13 |
Fold:Ribosomal protein L25-like
Superfamily:Ribosomal protein L25-like
Family:Gln-tRNA synthetase (GlnRS), C-terminal (anticodon-binding) domain |
| 29 | c3lk7A_ |
|
not modelled |
21.2 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: the crystal structure of udp-n-acetylmuramoylalanine-d-2 glutamate (murd) ligase from streptococcus agalactiae to3 1.5a
|
| 30 | d1wb9a2 |
|
not modelled |
21.1 |
44 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 31 | d2nzca1 |
|
not modelled |
20.9 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:TM1266-like |
| 32 | d1jr3d2 |
|
not modelled |
20.7 |
7 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 33 | d1i1ga2 |
|
not modelled |
19.9 |
15 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 34 | d4pfka_ |
|
not modelled |
19.9 |
26 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
| 35 | c2e1cA_ |
|
not modelled |
19.5 |
10 |
PDB header:transcription/dna
Chain: A: PDB Molecule:putative hth-type transcriptional regulator ph1519;
PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
|
| 36 | d2ev0a2 |
|
not modelled |
19.1 |
28 |
Fold:Iron-dependent repressor protein, dimerization domain
Superfamily:Iron-dependent repressor protein, dimerization domain
Family:Iron-dependent repressor protein, dimerization domain |
| 37 | d1fima_ |
|
not modelled |
19.0 |
7 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
| 38 | d1y14a_ |
|
not modelled |
18.7 |
21 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:RNA polymerase II subunit RBP4 (RpoF) |
| 39 | c1kyzC_ |
|
not modelled |
18.7 |
30 |
PDB header:transferase
Chain: C: PDB Molecule:caffeic acid 3-o-methyltransferase;
PDBTitle: crystal structure analysis of caffeic acid/5-hydroxyferulic2 acid 3/5-o-methyltransferase ferulic acid complex
|
| 40 | c3dx5A_ |
|
not modelled |
18.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 41 | c3canA_ |
|
not modelled |
18.4 |
22 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
| 42 | d1p9qc2 |
|
not modelled |
18.2 |
13 |
Fold:FYSH domain
Superfamily:FYSH domain
Family:Hypothetical protein AF0491, N-terminal domain |
| 43 | d1ul3a_ |
|
not modelled |
17.9 |
19 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 44 | d2r9ga1 |
|
not modelled |
17.8 |
13 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:MgsA/YrvN C-terminal domain-like |
| 45 | d2auna2 |
|
not modelled |
17.5 |
15 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:LD-carboxypeptidase A N-terminal domain-like |
| 46 | c2cveA_ |
|
not modelled |
17.2 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ttha1053;
PDBTitle: crystal structure of a conserved hypothetical protein tt1547 from2 thermus thermophilus hb8
|
| 47 | d1qmha1 |
|
not modelled |
16.7 |
29 |
Fold:Thioredoxin fold
Superfamily:RNA 3'-terminal phosphate cyclase, RPTC, insert domain
Family:RNA 3'-terminal phosphate cyclase, RPTC, insert domain |
| 48 | c1ewrA_ |
|
not modelled |
16.6 |
39 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna mismatch repair protein muts;
PDBTitle: crystal structure of taq muts
|
| 49 | d2cg4a2 |
|
not modelled |
16.4 |
13 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 50 | d3ctda1 |
|
not modelled |
16.2 |
13 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:MgsA/YrvN C-terminal domain-like |
| 51 | c2k6xA_ |
|
not modelled |
15.8 |
28 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpod;
PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
|
| 52 | c1d1rA_ |
|
not modelled |
15.5 |
21 |
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical 11.4 kd protein ycih in pyrf-osmb
PDBTitle: nmr solution structure of the product of the e. coli ycih2 gene.
|
| 53 | d1d1ra_ |
|
not modelled |
15.5 |
21 |
Fold:eIF1-like
Superfamily:eIF1-like
Family:eIF1-like |
| 54 | d1cc8a_ |
|
not modelled |
15.4 |
23 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 55 | c1zxxA_ |
|
not modelled |
15.4 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:6-phosphofructokinase;
PDBTitle: the crystal structure of phosphofructokinase from lactobacillus2 delbrueckii
|
| 56 | c1qr6A_ |
|
not modelled |
15.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malic enzyme 2;
PDBTitle: human mitochondrial nad(p)-dependent malic enzyme
|
| 57 | d3bgea1 |
|
not modelled |
15.3 |
13 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:MgsA/YrvN C-terminal domain-like |
| 58 | d1gd0a_ |
|
not modelled |
15.1 |
11 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
| 59 | c1o0sB_ |
|
not modelled |
15.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent malic enzyme;
PDBTitle: crystal structure of ascaris suum malic enzyme complexed with nadh
|
| 60 | c3thxB_ |
|
not modelled |
15.0 |
39 |
PDB header:dna binding protein/dna
Chain: B: PDB Molecule:dna mismatch repair protein msh3;
PDBTitle: human mutsbeta complexed with an idl of 3 bases (loop3) and adp
|
| 61 | c1ewqA_ |
|
not modelled |
14.7 |
39 |
PDB header:replication/dna
Chain: A: PDB Molecule:dna mismatch repair protein muts;
PDBTitle: crystal structure taq muts complexed with a heteroduplex2 dna at 2.2 a resolution
|
| 62 | d1pfka_ |
|
not modelled |
14.6 |
22 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
| 63 | c1b4aA_ |
|
not modelled |
14.3 |
15 |
PDB header:repressor
Chain: A: PDB Molecule:arginine repressor;
PDBTitle: structure of the arginine repressor from bacillus stearothermophilus
|
| 64 | d2gdga1 |
|
not modelled |
14.2 |
7 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
| 65 | d2vv5a2 |
|
not modelled |
14.0 |
11 |
Fold:Ferredoxin-like
Superfamily:Mechanosensitive channel protein MscS (YggB), C-terminal domain
Family:Mechanosensitive channel protein MscS (YggB), C-terminal domain |
| 66 | c2o8dB_ |
|
not modelled |
14.0 |
35 |
PDB header:dna binding protein/dna
Chain: B: PDB Molecule:dna mismatch repair protein msh6;
PDBTitle: human mutsalpha (msh2/msh6) bound to adp and a g du mispair
|
| 67 | c3ctdB_ |
|
not modelled |
13.9 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative atpase, aaa family;
PDBTitle: crystal structure of a putative aaa family atpase from2 prochlorococcus marinus subsp. pastoris
|
| 68 | c3iwcD_ |
|
not modelled |
13.7 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:s-adenosylmethionine decarboxylase;
PDBTitle: t. maritima adometdc complex with s-adenosylmethionine2 methyl ester
|
| 69 | c3fybA_ |
|
not modelled |
13.6 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein of unknown function (duf1244);
PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
|
| 70 | c3b64A_ |
|
not modelled |
13.5 |
14 |
PDB header:cytokine
Chain: A: PDB Molecule:macrophage migration inhibitory factor-like
PDBTitle: macrophage migration inhibitory factor (mif) from2 /leishmania major
|
| 71 | d2fgca1 |
|
not modelled |
13.4 |
20 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 72 | d2f06a1 |
|
not modelled |
13.1 |
22 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 73 | d2o35a1 |
|
not modelled |
13.0 |
29 |
Fold:SMc04008-like
Superfamily:SMc04008-like
Family:SMc04008-like |
| 74 | c2o35A_ |
|
not modelled |
13.0 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein duf1244;
PDBTitle: protein of unknown function (duf1244) from sinorhizobium meliloti
|
| 75 | c1i1gA_ |
|
not modelled |
12.7 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator lrpa;
PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
|
| 76 | c3ereD_ |
|
not modelled |
12.5 |
18 |
PDB header:dna binding protein/dna
Chain: D: PDB Molecule:arginine repressor;
PDBTitle: crystal structure of the arginine repressor protein from mycobacterium2 tuberculosis in complex with the dna operator
|
| 77 | d1xtpa_ |
|
not modelled |
12.5 |
24 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:AD-003 protein-like |
| 78 | c2pc6C_ |
|
not modelled |
12.4 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
| 79 | d1pkxa1 |
|
not modelled |
12.2 |
14 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 80 | c2e1aD_ |
|
not modelled |
12.1 |
13 |
PDB header:transcription
Chain: D: PDB Molecule:75aa long hypothetical regulatory protein asnc;
PDBTitle: crystal structure of ffrp-dm1
|
| 81 | c3k85B_ |
|
not modelled |
11.9 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:d-glycero-d-manno-heptose 1-phosphate kinase;
PDBTitle: crystal structure of a d-glycero-d-manno-heptose 1-phosphate2 kinase from bacteriodes thetaiotaomicron
|
| 82 | d2fxaa1 |
|
not modelled |
11.9 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 83 | c3ia1A_ |
|
not modelled |
11.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thio-disulfide isomerase/thioredoxin;
PDBTitle: crystal structure of thio-disulfide isomerase from thermus2 thermophilus
|
| 84 | d2adca1 |
|
not modelled |
11.9 |
14 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 85 | c2w2uA_ |
|
not modelled |
11.9 |
25 |
PDB header:hydrolase/transport
Chain: A: PDB Molecule:hypothetical p60 katanin;
PDBTitle: structural insight into the interaction between archaeal2 escrt-iii and aaa-atpase
|
| 86 | c2qt7B_ |
|
not modelled |
11.8 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:receptor-type tyrosine-protein phosphatase-like
PDBTitle: crystallographic structure of the mature ectodomain of the2 human receptor-type protein-tyrosine phosphatase ia-2 at3 1.30 angstroms
|
| 87 | c2i38A_ |
|
not modelled |
11.7 |
17 |
PDB header:rna binding protein/chimera
Chain: A: PDB Molecule:fusion protein consists of immunoglobin g-
PDBTitle: solution structure of the rrm of srp20
|
| 88 | d2u1aa_ |
|
not modelled |
11.6 |
10 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 89 | c2cb4D_ |
|
not modelled |
11.5 |
40 |
PDB header:toxin
Chain: D: PDB Molecule:mosquitocidal toxin;
PDBTitle: crystal structure of the catalytic domain of the2 mosquitocidal toxin from bacillus sphaericus, mutant e197q
|
| 90 | d1vr7a1 |
|
not modelled |
11.4 |
20 |
Fold:S-adenosylmethionine decarboxylase
Superfamily:S-adenosylmethionine decarboxylase
Family:Bacterial S-adenosylmethionine decarboxylase |
| 91 | c3p6lA_ |
|
not modelled |
11.3 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
| 92 | d1fp1d2 |
|
not modelled |
11.3 |
27 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 93 | c3jz3B_ |
|
not modelled |
11.2 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:sensor protein qsec;
PDBTitle: structure of the cytoplasmic segment of histidine kinase qsec
|
| 94 | c2d9oA_ |
|
not modelled |
11.1 |
8 |
PDB header:rna binding protein
Chain: A: PDB Molecule:dnaj (hsp40) homolog, subfamily c, member 17;
PDBTitle: solution structure of rna binding domain in hypothetical2 protein flj10634
|
| 95 | c2p6tH_ |
|
not modelled |
11.1 |
16 |
PDB header:transcription
Chain: H: PDB Molecule:transcriptional regulator, lrp/asnc family;
PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
|
| 96 | c2vxcA_ |
|
not modelled |
11.0 |
12 |
PDB header:cell cycle
Chain: A: PDB Molecule:dna repair protein rhp9;
PDBTitle: structure of the crb2-brct2 domain complex with2 phosphopeptide.
|
| 97 | d1mwza_ |
|
not modelled |
10.9 |
17 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 98 | c2o8bA_ |
|
not modelled |
10.9 |
48 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:dna mismatch repair protein msh2;
PDBTitle: human mutsalpha (msh2/msh6) bound to adp and a g t mispair
|
| 99 | c1wbdA_ |
|
not modelled |
10.8 |
39 |
PDB header:dna-binding
Chain: A: PDB Molecule:dna mismatch repair protein muts;
PDBTitle: crystal structure of e. coli dna mismatch repair enzyme2 muts, e38q mutant, in complex with a g.t mismatch
|