| 1 | c3bdkB_
|
|
 |
100.0 |
45 |
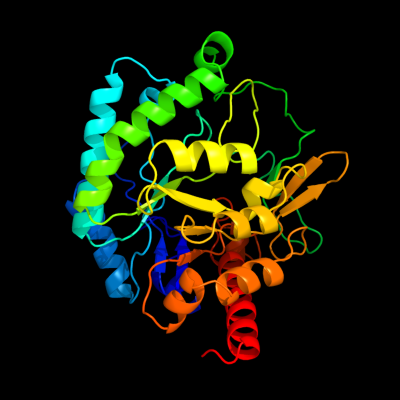
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
| 2 | d1tz9a_
|
|
 |
100.0 |
41 |
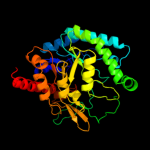
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:UxuA-like |
| 3 | c2hk1D_
|
|
 |
99.9 |
18 |
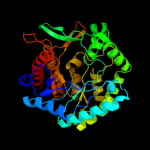
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
| 4 | d1i60a_
|
|
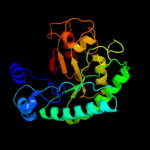 |
99.9 |
16 |
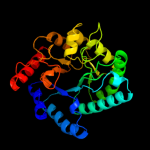
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 5 | c3kwsB_
|
|
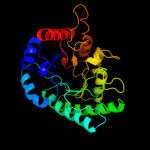 |
99.9 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
| 6 | c2ou4C_
|
|
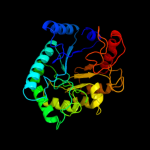 |
99.9 |
19 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
| 7 | c3qxbB_
|
|
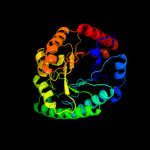 |
99.9 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:putative xylose isomerase;
PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
|
| 8 | c3cnyA_
|
|
 |
99.9 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
| 9 | d1k77a_
|
|
 |
99.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Hypothetical protein YgbM (EC1530) |
| 10 | c3obeB_
|
|
 |
99.9 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
|
| 11 | c3lmzA_
|
|
 |
99.9 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase. (yp_001305105.1) from2 parabacteroides distasonis atcc 8503 at 1.44 a resolution
|
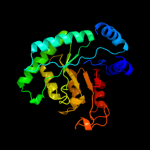
| 12 | c3dx5A_
|
|
 |
99.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
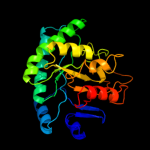
| 13 | c3ju2A_
|
|
 |
99.9 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein smc04130;
PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
|
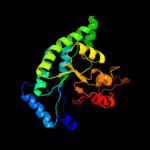
| 14 | c2zdsB_
|
|
 |
99.9 |
18 |
PDB header:dna binding protein
Chain: B: PDB Molecule:putative dna-binding protein;
PDBTitle: crystal structure of sco6571 from streptomyces coelicolor2 a3(2)
|
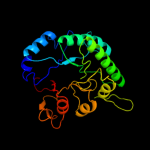
| 15 | c3l23A_
|
|
 |
99.8 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
|
| 16 | c3ngfA_
|
|
 |
99.8 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family 2;
PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
|
| 17 | c3p6lA_
|
|
 |
99.8 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
| 18 | c2qw5B_
|
|
 |
99.8 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase-like tim barrel;
PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
|
| 19 | c3ktcB_
|
|
 |
99.8 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
|
| 20 | c2zvrA_
|
|
 |
99.8 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related2 protein from thermotoga maritima
|
| 21 | d2g0wa1 |
|
not modelled |
99.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 22 | c3cqkB_ |
|
not modelled |
99.8 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 23 | d1xp3a1 |
|
not modelled |
99.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
| 24 | d2q02a1 |
|
not modelled |
99.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 25 | d1yx1a1 |
|
not modelled |
99.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
| 26 | d1qtwa_ |
|
not modelled |
99.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
| 27 | d1bxca_ |
|
not modelled |
99.7 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 28 | d2glka1 |
|
not modelled |
99.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 29 | d1muwa_ |
|
not modelled |
99.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 30 | d1bxba_ |
|
not modelled |
99.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 31 | c2x7vA_ |
|
not modelled |
99.6 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
| 32 | d1qt1a_ |
|
not modelled |
99.5 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 33 | d1xima_ |
|
not modelled |
99.5 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 34 | c3aamA_ |
|
not modelled |
99.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease iv;
PDBTitle: crystal structure of endonuclease iv from thermus thermophilus hb8
|
| 35 | d1xlma_ |
|
not modelled |
99.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 36 | c2i56A_ |
|
not modelled |
98.7 |
16 |
PDB header:isomerase, metal-binding protein
Chain: A: PDB Molecule:l-rhamnose isomerase;
PDBTitle: crystal structure of l-rhamnose isomerase from pseudomonas2 stutzeri with l-rhamnose
|
| 37 | c3bzjA_ |
|
not modelled |
97.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:uv endonuclease;
PDBTitle: uvde k229l
|
| 38 | d1a0ea_ |
|
not modelled |
96.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 39 | d1a0da_ |
|
not modelled |
96.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 40 | d1a0ca_ |
|
not modelled |
95.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 41 | c3p14C_ |
|
not modelled |
94.8 |
21 |
PDB header:isomerase
Chain: C: PDB Molecule:l-rhamnose isomerase;
PDBTitle: crystal structure of l-rhamnose isomerase with a novel high thermo-2 stability from bacillus halodurans
|
| 42 | c3bwwA_ |
|
not modelled |
92.3 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:protein of unknown function duf692/cog3220;
PDBTitle: crystal structure of a duf692 family protein (hs_1138) from2 haemophilus somnus 129pt at 2.20 a resolution
|
| 43 | d1ob0a2 |
|
not modelled |
86.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 44 | c1sr9A_ |
|
not modelled |
83.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of leua from mycobacterium tuberculosis
|
| 45 | c3atyA_ |
|
not modelled |
83.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prostaglandin f2a synthase;
PDBTitle: crystal structure of tcoye
|
| 46 | d1vgga_ |
|
not modelled |
81.7 |
25 |
Fold:Ta1353-like
Superfamily:Ta1353-like
Family:Ta1353-like |
| 47 | c2ekmC_ |
|
not modelled |
80.1 |
22 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein st1511;
PDBTitle: structure of st1219 protein from sulfolobus tokodaii
|
| 48 | c3hpxB_ |
|
not modelled |
78.2 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of mycobacterium tuberculosis leua active site2 domain 1-425 (truncation mutant delta:426-644)
|
| 49 | c2p10D_ |
|
not modelled |
77.8 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 50 | c2gl0A_ |
|
not modelled |
77.5 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: structure of pae2307 in complex with adenosine
|
| 51 | c2xfyA_ |
|
not modelled |
77.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-amylase;
PDBTitle: crystal structure of barley beta-amylase complexed with2 alpha-cyclodextrin
|
| 52 | d1olta_ |
|
not modelled |
77.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
| 53 | d1fa2a_ |
|
not modelled |
77.3 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 54 | c2c3zA_ |
|
not modelled |
77.3 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
| 55 | d1wdpa1 |
|
not modelled |
77.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 56 | d1mxga2 |
|
not modelled |
76.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 57 | d1to3a_ |
|
not modelled |
75.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 58 | c3iacA_ |
|
not modelled |
74.6 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:glucuronate isomerase;
PDBTitle: 2.2 angstrom crystal structure of glucuronate isomerase from2 salmonella typhimurium.
|
| 59 | d1b1ya_ |
|
not modelled |
73.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 60 | d2p10a1 |
|
not modelled |
72.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Mll9387-like |
| 61 | d1avaa2 |
|
not modelled |
71.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 62 | d1r30a_ |
|
not modelled |
71.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
| 63 | c1r30A_ |
|
not modelled |
71.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
|
| 64 | d1rlha_ |
|
not modelled |
70.9 |
25 |
Fold:Ta1353-like
Superfamily:Ta1353-like
Family:Ta1353-like |
| 65 | d1hvxa2 |
|
not modelled |
70.5 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 66 | c2vg2C_ |
|
not modelled |
65.4 |
12 |
PDB header:transferase
Chain: C: PDB Molecule:undecaprenyl pyrophosphate synthetase;
PDBTitle: rv2361 with ipp
|
| 67 | d1hx0a2 |
|
not modelled |
63.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 68 | d2gjpa2 |
|
not modelled |
63.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 69 | c3eb2A_ |
|
not modelled |
62.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 70 | d1x7fa2 |
|
not modelled |
62.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Outer surface protein, N-terminal domain |
| 71 | d1tj1a2 |
|
not modelled |
59.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FAD-linked oxidoreductase
Family:Proline dehydrohenase domain of bifunctional PutA protein |
| 72 | d1vema2 |
|
not modelled |
59.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 73 | d3dhpa2 |
|
not modelled |
58.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 74 | c2vc7A_ |
|
not modelled |
57.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:aryldialkylphosphatase;
PDBTitle: structural basis for natural lactonase and promiscuous2 phosphotriesterase activities
|
| 75 | d1i4na_ |
|
not modelled |
55.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 76 | c3blpX_ |
|
not modelled |
55.5 |
19 |
PDB header:hydrolase
Chain: X: PDB Molecule:alpha-amylase 1;
PDBTitle: role of aromatic residues in human salivary alpha-amylase
|
| 77 | c2yusA_ |
|
not modelled |
55.0 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:swi/snf-related matrix-associated actin-
PDBTitle: solution structure of the sant domain of human swi/snf-2 related matrix-associated actin-dependent regulator of3 chromatin subfamily c member 1
|
| 78 | d1ud2a2 |
|
not modelled |
53.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 79 | c3cixA_ |
|
not modelled |
52.6 |
19 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
| 80 | d1eg7a_ |
|
not modelled |
51.8 |
24 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 81 | c1tdjA_ |
|
not modelled |
51.0 |
13 |
PDB header:allostery
Chain: A: PDB Molecule:biosynthetic threonine deaminase;
PDBTitle: threonine deaminase (biosynthetic) from e. coli
|
| 82 | d2cu7a1 |
|
not modelled |
49.2 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 83 | d1g5aa2 |
|
not modelled |
48.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 84 | c2ehhE_ |
|
not modelled |
47.8 |
17 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 85 | c2yumA_ |
|
not modelled |
47.2 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger zz-type-containing protein 3;
PDBTitle: solution structure of the myb-like dna-binding domain of2 human zzz3 protein
|
| 86 | c3dhuC_ |
|
not modelled |
46.8 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure of an alpha-amylase from lactobacillus2 plantarum
|
| 87 | d1phza1 |
|
not modelled |
46.8 |
20 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 88 | c2d16B_ |
|
not modelled |
45.8 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph1918;
PDBTitle: crystal structure of ph1918 protein from pyrococcus horikoshii ot3
|
| 89 | d1e9yb2 |
|
not modelled |
45.7 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:alpha-subunit of urease, catalytic domain |
| 90 | d1d8wa_ |
|
not modelled |
45.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:L-rhamnose isomerase |
| 91 | c1x7fA_ |
|
not modelled |
45.2 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:outer surface protein;
PDBTitle: crystal structure of an uncharacterized b. cereus protein
|
| 92 | d1ujpa_ |
|
not modelled |
43.7 |
36 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 93 | d1ejxc2 |
|
not modelled |
43.3 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:alpha-subunit of urease, catalytic domain |
| 94 | c3pnuA_ |
|
not modelled |
42.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:dihydroorotase;
PDBTitle: 2.4 angstrom crystal structure of dihydroorotase (pyrc) from2 campylobacter jejuni.
|
| 95 | d1ht6a2 |
|
not modelled |
41.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 96 | c1rr2A_ |
|
not modelled |
41.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 97 | c3f6tA_ |
|
not modelled |
41.0 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate aminotransferase;
PDBTitle: crystal structure of aspartate aminotransferase (e.c. 2.6.1.1)2 (yp_194538.1) from lactobacillus acidophilus ncfm at 2.15 a3 resolution
|
| 98 | d2cbia2 |
|
not modelled |
40.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:alpha-D-glucuronidase/Hyaluronidase catalytic domain |
| 99 | c1ud8A_ |
|
not modelled |
40.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:amylase;
PDBTitle: crystal structure of amyk38 with lithium ion
|
| 100 | d1e43a2 |
|
not modelled |
40.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 101 | d1foba_ |
|
not modelled |
40.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 102 | d1hjqa_ |
|
not modelled |
40.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 103 | c2o8vA_ |
|
not modelled |
39.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: paps reductase in a covalent complex with thioredoxin c35a
|
| 104 | d2aaaa2 |
|
not modelled |
39.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 105 | c2zy3A_ |
|
not modelled |
39.6 |
6 |
PDB header:lyase
Chain: A: PDB Molecule:l-aspartate beta-decarboxylase;
PDBTitle: dodecameric l-aspartate beta-decarboxylase
|
| 106 | c2yxgD_ |
|
not modelled |
39.5 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 107 | d1m7xa3 |
|
not modelled |
39.3 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 108 | c2p0oA_ |
|
not modelled |
39.1 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein duf871;
PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
|
| 109 | d1piia2 |
|
not modelled |
39.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 110 | c3k13A_ |
|
not modelled |
39.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 111 | c3t7vA_ |
|
not modelled |
39.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
| 112 | d1j5ta_ |
|
not modelled |
39.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 113 | d1vr3a1 |
|
not modelled |
38.2 |
24 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Acireductone dioxygenase |
| 114 | c2wcsA_ |
|
not modelled |
38.0 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha amylase, catalytic region;
PDBTitle: crystal structure of debranching enzyme from nostoc2 punctiforme (npde)
|
| 115 | d2bhua3 |
|
not modelled |
37.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 116 | c3amlA_ |
|
not modelled |
37.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:os06g0726400 protein;
PDBTitle: structure of the starch branching enzyme i (bei) from oryza sativa l
|
| 117 | d1j5sa_ |
|
not modelled |
37.0 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Uronate isomerase-like |
| 118 | c2e8yA_ |
|
not modelled |
36.8 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:amyx protein;
PDBTitle: crystal structure of pullulanase type i from bacillus subtilis str.2 168
|
| 119 | d1jaea2 |
|
not modelled |
36.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 120 | c1zlpA_ |
|
not modelled |
36.4 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|