| 1 | c1w25B_ |
|
 |
100.0 |
32 |
PDB header:signaling protein
Chain: B: PDB Molecule:stalked-cell differentiation controlling protein;
PDBTitle: response regulator pled in complex with c-digmp
|
| 2 | c3ezuA_ |
|
 |
100.0 |
24 |
PDB header:signaling protein
Chain: A: PDB Molecule:ggdef domain protein;
PDBTitle: crystal structure of multidomain protein of unknown function with2 ggdef-domain (np_951600.1) from geobacter sulfurreducens at 1.95 a3 resolution
|
| 3 | c3breA_ |
|
 |
100.0 |
26 |
PDB header:signaling protein
Chain: A: PDB Molecule:probable two-component response regulator;
PDBTitle: crystal structure of p.aeruginosa pa3702
|
| 4 | c3i5aA_ |
|
 |
100.0 |
26 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator/ggdef domain protein;
PDBTitle: crystal structure of full-length wpsr from pseudomonas syringae
|
| 5 | c3qyyB_ |
|
 |
100.0 |
27 |
PDB header:signaling protein/inhibitor
Chain: B: PDB Molecule:response regulator;
PDBTitle: a novel interaction mode between a microbial ggdef domain and the bis-2 (3, 5 )-cyclic di-gmp
|
| 6 | c3i5cA_ |
|
 |
100.0 |
30 |
PDB header:signaling protein
Chain: A: PDB Molecule:fusion of general control protein gcn4 and wspr response
PDBTitle: crystal structure of a fusion protein containing the leucine zipper of2 gcn4 and the ggdef domain of wspr from pseudomonas aeruginosa
|
| 7 | c3i5bA_ |
|
 |
100.0 |
30 |
PDB header:signaling protein
Chain: A: PDB Molecule:wspr response regulator;
PDBTitle: crystal structure of the isolated ggdef domain of wpsr from2 pseudomonas aeruginosa
|
| 8 | c3hvaA_ |
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:protein fimx;
PDBTitle: crystal structure of fimx ggdef domain from pseudomonas2 aeruginosa
|
| 9 | c3iclA_ |
|
 |
100.0 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:eal/ggdef domain protein;
PDBTitle: x-ray structure of protein (eal/ggdef domain protein) from2 m.capsulatus, northeast structural genomics consortium3 target mcr174c
|
| 10 | c3ignA_ |
|
 |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:diguanylate cyclase;
PDBTitle: crystal structure of the ggdef domain from marinobacter2 aquaeolei diguanylate cyclase complexed with c-di-gmp -3 northeast structural genomics consortium target mqr89a
|
| 11 | d1w25a3 |
|
 |
100.0 |
38 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:GGDEF domain |
| 12 | c3mtkA_ |
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:diguanylate cyclase/phosphodiesterase;
PDBTitle: x-ray structure of diguanylate cyclase/phosphodiesterase from2 caldicellulosiruptor saccharolyticus, northeast structural genomics3 consortium target clr27c
|
| 13 | c3pjwA_ |
|
 |
100.0 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:cyclic dimeric gmp binding protein;
PDBTitle: structure of pseudomonas fluorescence lapd ggdef-eal dual domain, i23
|
| 14 | c3hvwA_ |
|
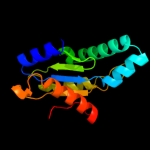 |
100.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:diguanylate-cyclase (dgc);
PDBTitle: crystal structure of the ggdef domain of the pa2567 protein2 from pseudomonas aeruginosa, northeast structural genomics3 consortium target par365c
|
| 15 | c3gfzB_ |
|
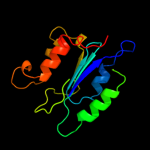 |
99.8 |
13 |
PDB header:hydrolase, signaling protein
Chain: B: PDB Molecule:klebsiella pneumoniae blrp1;
PDBTitle: klebsiella pneumoniae blrp1 ph 6 manganese/cy-digmp complex
|
| 16 | c3p7nB_ |
|
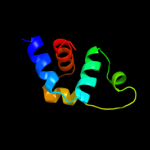 |
98.9 |
8 |
PDB header:dna binding protein
Chain: B: PDB Molecule:sensor histidine kinase;
PDBTitle: crystal structure of light activated transcription factor el222 from2 erythrobacter litoralis
|
| 17 | c2qv6D_ |
|
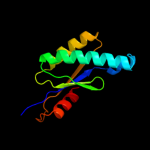 |
97.7 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase iii;
PDBTitle: gtp cyclohydrolase iii from m. jannaschii (mj0145)2 complexed with gtp and metal ions
|
| 18 | d1fx2a_ |
|
 |
94.7 |
16 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
| 19 | c1wc6B_ |
|
 |
93.6 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:adenylate cyclase;
PDBTitle: soluble adenylyl cyclase cyac from s. platensis in complex2 with rp-atpalphas in presence of bicarbonate
|
| 20 | c3mr7B_ |
|
 |
92.7 |
9 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenylate/guanylate cyclase/hydrolase, alpha/beta fold
PDBTitle: crystal structure of adenylate/guanylate cyclase/hydrolase from2 silicibacter pomeroyi
|
| 21 | c1cjkA_ |
|
not modelled |
91.6 |
8 |
PDB header:lyase/lyase/signaling protein
Chain: A: PDB Molecule:adenylate cyclase, type v;
PDBTitle: complex of gs-alpha with the catalytic domains of mammalian adenylyl2 cyclase: complex with adenosine 5'-(alpha thio)-triphosphate (rp),3 mg, and mn
|
| 22 | c2w01C_ |
|
not modelled |
91.4 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:adenylate cyclase;
PDBTitle: crystal structure of the guanylyl cyclase cya2
|
| 23 | d1wc1a_ |
|
not modelled |
91.0 |
15 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
| 24 | c1s97D_ |
|
not modelled |
90.2 |
15 |
PDB header:transferase/dna
Chain: D: PDB Molecule:dna polymerase iv;
PDBTitle: dpo4 with gt mismatch
|
| 25 | c2aq4A_ |
|
not modelled |
89.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:dna repair protein rev1;
PDBTitle: ternary complex of the catalytic core of rev1 with dna and dctp.
|
| 26 | c1ybuA_ |
|
not modelled |
88.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipj;
PDBTitle: mycobacterium tuberculosis adenylyl cyclase rv1900c chd, in complex2 with a substrate analog.
|
| 27 | d1azsa_ |
|
not modelled |
88.2 |
8 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
| 28 | d1fx4a_ |
|
not modelled |
88.0 |
14 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
| 29 | c3uvjC_ |
|
not modelled |
87.8 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:guanylate cyclase soluble subunit alpha-3;
PDBTitle: crystal structure of the catalytic domain of the heterodimeric human2 soluble guanylate cyclase 1.
|
| 30 | c3gqcB_ |
|
not modelled |
87.7 |
19 |
PDB header:transferase/dna
Chain: B: PDB Molecule:dna repair protein rev1;
PDBTitle: structure of human rev1-dna-dntp ternary complex
|
| 31 | c1k1qA_ |
|
not modelled |
84.7 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:dbh protein;
PDBTitle: crystal structure of a dinb family error prone dna2 polymerase from sulfolobus solfataricus
|
| 32 | d1k1sa2 |
|
not modelled |
83.9 |
15 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 33 | c2r8kB_ |
|
not modelled |
80.0 |
24 |
PDB header:replication, transferase/dna
Chain: B: PDB Molecule:dna polymerase eta;
PDBTitle: structure of the eukaryotic dna polymerase eta in complex with 1,2-2 d(gpg)-cisplatin containing dna
|
| 34 | c3onqB_ |
|
not modelled |
79.7 |
4 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:regulator of polyketide synthase expression;
PDBTitle: crystal structure of regulator of polyketide synthase expression2 bad_0249 from bifidobacterium adolescentis
|
| 35 | d1jx4a2 |
|
not modelled |
78.6 |
13 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 36 | d1im4a_ |
|
not modelled |
76.2 |
14 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 37 | c1jihA_ |
|
not modelled |
74.9 |
24 |
PDB header:translation
Chain: A: PDB Molecule:dna polymerase eta;
PDBTitle: yeast dna polymerase eta
|
| 38 | c3et6A_ |
|
not modelled |
72.6 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:soluble guanylyl cyclase beta;
PDBTitle: the crystal structure of the catalytic domain of a eukaryotic2 guanylate cyclase
|
| 39 | c1yk9A_ |
|
not modelled |
72.3 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:adenylate cyclase;
PDBTitle: crystal structure of a mutant form of the mycobacterial2 adenylyl cyclase rv1625c
|
| 40 | d1jiha2 |
|
not modelled |
65.7 |
23 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 41 | c1t3nB_ |
|
not modelled |
65.3 |
34 |
PDB header:replication/dna
Chain: B: PDB Molecule:polymerase (dna directed) iota;
PDBTitle: structure of the catalytic core of dna polymerase iota in2 complex with dna and dttp
|
| 42 | c2fllA_ |
|
not modelled |
63.1 |
31 |
PDB header:replication/dna
Chain: A: PDB Molecule:dna polymerase iota;
PDBTitle: ternary complex of human dna polymerase iota with dna and dttp
|
| 43 | c1t94B_ |
|
not modelled |
62.2 |
24 |
PDB header:replication
Chain: B: PDB Molecule:polymerase (dna directed) kappa;
PDBTitle: crystal structure of the catalytic core of human dna2 polymerase kappa
|
| 44 | c2oh2B_ |
|
not modelled |
62.0 |
21 |
PDB header:transferase/dna
Chain: B: PDB Molecule:dna polymerase kappa;
PDBTitle: ternary complex of human dna polymerase
|
| 45 | c3mr2A_ |
|
not modelled |
60.4 |
23 |
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase eta;
PDBTitle: human dna polymerase eta in complex with normal dna and incoming2 nucleotide (nrm)
|
| 46 | c1y10C_ |
|
not modelled |
56.1 |
9 |
PDB header:lyase
Chain: C: PDB Molecule:hypothetical protein rv1264/mt1302;
PDBTitle: mycobacterial adenylyl cyclase rv1264, holoenzyme, inhibited state
|
| 47 | c2wz1B_ |
|
not modelled |
44.7 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:guanylate cyclase soluble subunit beta-1;
PDBTitle: structure of the catalytic domain of human soluble2 guanylate cyclase 1 beta 3.
|
| 48 | d1t94a2 |
|
not modelled |
30.8 |
23 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 49 | c1vloA_ |
|
not modelled |
29.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of aminomethyltransferase (t protein;2 tetrahydrofolate-dependent) of glycine cleavage system (np417381)3 from escherichia coli k12 at 1.70 a resolution
|
| 50 | d1yhta1 |
|
not modelled |
28.5 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 51 | c3hf3A_ |
|
not modelled |
27.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 52 | c3mpbA_ |
|
not modelled |
23.7 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar isomerase;
PDBTitle: z5688 from e. coli o157:h7 bound to fructose
|
| 53 | c2h90A_ |
|
not modelled |
21.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
| 54 | d1ps9a1 |
|
not modelled |
19.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 55 | c1rgsA_ |
|
not modelled |
19.7 |
24 |
PDB header:kinase
Chain: A: PDB Molecule:camp dependent protein kinase;
PDBTitle: regulatory subunit of camp dependent protein kinase
|
| 56 | c2byvE_ |
|
not modelled |
18.9 |
15 |
PDB header:regulation
Chain: E: PDB Molecule:rap guanine nucleotide exchange factor 4;
PDBTitle: structure of the camp responsive exchange factor epac2 in2 its auto-inhibited state
|
| 57 | d1mo1a_ |
|
not modelled |
18.8 |
32 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 58 | d1ptfa_ |
|
not modelled |
17.3 |
14 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 59 | c1n13J_ |
|
not modelled |
17.0 |
17 |
PDB header:lyase
Chain: J: PDB Molecule:pyruvoyl-dependent arginine decarboxylase alpha
PDBTitle: the crystal structure of pyruvoyl-dependent arginine2 decarboxylase from methanococcus jannashii
|
| 60 | c3in6A_ |
|
not modelled |
16.8 |
42 |
PDB header:flavoprotein
Chain: A: PDB Molecule:fmn-binding protein;
PDBTitle: crystal structure of a fmn-binding protein (swol_0183) from2 syntrophomonas wolfei subsp. wolfei at 2.12 a resolution
|
| 61 | d1cm3a_ |
|
not modelled |
16.5 |
12 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 62 | d1zeta2 |
|
not modelled |
16.4 |
41 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 63 | d3c9fa2 |
|
not modelled |
15.8 |
24 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 64 | c3ihsB_ |
|
not modelled |
15.6 |
28 |
PDB header:transport protein
Chain: B: PDB Molecule:phosphocarrier protein hpr;
PDBTitle: crystal structure of a phosphocarrier protein hpr from2 bacillus anthracis str. ames
|
| 65 | c1o7fA_ |
|
not modelled |
15.1 |
14 |
PDB header:regulation
Chain: A: PDB Molecule:camp-dependent rap1 guanine-nucleotide exchange
PDBTitle: crystal structure of the regulatory domain of epac2
|
| 66 | d1zvvj1 |
|
not modelled |
13.6 |
32 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 67 | c3le1B_ |
|
not modelled |
12.5 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:phosphotransferase system, hpr-related proteins;
PDBTitle: crystal structure of apohpr monomer from thermoanaerobacter2 tengcongensis
|
| 68 | c3k30B_ |
|
not modelled |
12.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
| 69 | d1qr5a_ |
|
not modelled |
11.9 |
7 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 70 | c3shrA_ |
|
not modelled |
11.8 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:cgmp-dependent protein kinase 1;
PDBTitle: crystal structure of cgmp-dependent protein kinase reveals novel site2 of interchain communication
|
| 71 | c2i34B_ |
|
not modelled |
11.8 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:acid phosphatase;
PDBTitle: the crystal structure of class c acid phosphatase from bacillus2 anthracis with tungstate bound
|
| 72 | c3rbuA_ |
|
not modelled |
11.7 |
19 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:glutamate carboxypeptidase 2;
PDBTitle: n-terminally avitev-tagged human glutamate carboxypeptidase ii in2 complex with 2-pmpa
|
| 73 | d1k3xa1 |
|
not modelled |
11.4 |
30 |
Fold:S13-like H2TH domain
Superfamily:S13-like H2TH domain
Family:Middle domain of MutM-like DNA repair proteins |
| 74 | c2ootA_ |
|
not modelled |
10.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:glutamate carboxypeptidase 2;
PDBTitle: a high resolution structure of ligand-free human glutamate2 carboxypeptidase ii
|
| 75 | d1ka5a_ |
|
not modelled |
10.7 |
8 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 76 | c3c9fB_ |
|
not modelled |
10.7 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase from candida albicans sc5314
|
| 77 | d1wosa2 |
|
not modelled |
10.5 |
21 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
| 78 | d1nowa1 |
|
not modelled |
10.5 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 79 | d1qbaa3 |
|
not modelled |
10.4 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 80 | c3of1A_ |
|
not modelled |
10.3 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:camp-dependent protein kinase regulatory subunit;
PDBTitle: crystal structure of bcy1, the yeast regulatory subunit of pka
|
| 81 | c1cx8F_ |
|
not modelled |
10.2 |
16 |
PDB header:metal transport
Chain: F: PDB Molecule:transferrin receptor protein;
PDBTitle: crytal structure of the ectodomain of human transferrin receptor
|
| 82 | d1vr6a1 |
|
not modelled |
9.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 83 | c3fgeA_ |
|
not modelled |
9.7 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative flavin reductase with split barrel domain;
PDBTitle: crystal structure of putative flavin reductase with split barrel2 domain (yp_750721.1) from shewanella frigidimarina ncimb 400 at 1.743 a resolution
|
| 84 | d2o5aa1 |
|
not modelled |
9.7 |
18 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Iojap/YbeB-like |
| 85 | d1cgme_ |
|
not modelled |
9.6 |
19 |
Fold:Four-helical up-and-down bundle
Superfamily:TMV-like viral coat proteins
Family:TMV-like viral coat proteins |
| 86 | d1tdza1 |
|
not modelled |
9.6 |
40 |
Fold:S13-like H2TH domain
Superfamily:S13-like H2TH domain
Family:Middle domain of MutM-like DNA repair proteins |
| 87 | c1worA_ |
|
not modelled |
9.5 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of t-protein of the glycine cleavage2 system
|
| 88 | d1ei7a_ |
|
not modelled |
9.4 |
36 |
Fold:Four-helical up-and-down bundle
Superfamily:TMV-like viral coat proteins
Family:TMV-like viral coat proteins |
| 89 | c3kmlB_ |
|
not modelled |
9.1 |
36 |
PDB header:viral protein
Chain: B: PDB Molecule:coat protein;
PDBTitle: circular permutant of the tobacco mosaic virus
|
| 90 | c3iyuY_ |
|
not modelled |
9.1 |
19 |
PDB header:virus
Chain: Y: PDB Molecule:outer capsid protein vp4;
PDBTitle: atomic model of an infectious rotavirus particle
|
| 91 | c1ps9A_ |
|
not modelled |
9.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 92 | c1qbaA_ |
|
not modelled |
9.0 |
33 |
PDB header:glycosyl hydrolase
Chain: A: PDB Molecule:chitobiase;
PDBTitle: bacterial chitobiase, glycosyl hydrolase family 20
|
| 93 | d1ee8a1 |
|
not modelled |
8.8 |
40 |
Fold:S13-like H2TH domain
Superfamily:S13-like H2TH domain
Family:Middle domain of MutM-like DNA repair proteins |
| 94 | c2jroA_ |
|
not modelled |
8.6 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of so0334 from shewanella oneidensis. northeast2 structural genomics target sor75
|
| 95 | d1vh8a_ |
|
not modelled |
8.5 |
18 |
Fold:Bacillus chorismate mutase-like
Superfamily:IpsF-like
Family:IpsF-like |
| 96 | c2wk1A_ |
|
not modelled |
8.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:novp;
PDBTitle: structure of the o-methyltransferase novp
|
| 97 | d1k82a1 |
|
not modelled |
8.1 |
30 |
Fold:S13-like H2TH domain
Superfamily:S13-like H2TH domain
Family:Middle domain of MutM-like DNA repair proteins |
| 98 | c3gveB_ |
|
not modelled |
8.0 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:yfkn protein;
PDBTitle: crystal structure of calcineurin-like phosphoesterase yfkn from2 bacillus subtilis
|
| 99 | c2ylaA_ |
|
not modelled |
8.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|

