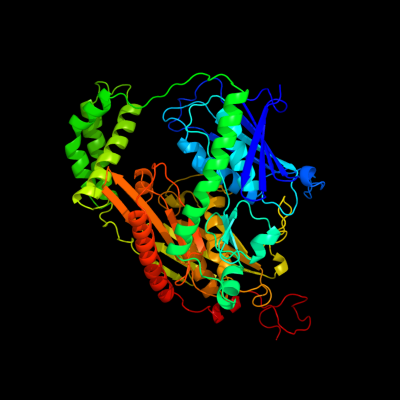
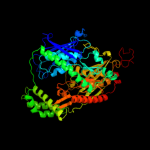
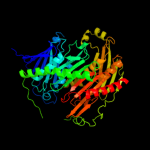
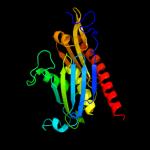
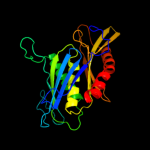
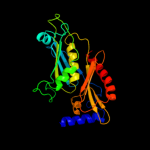
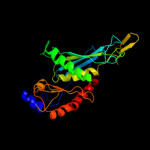
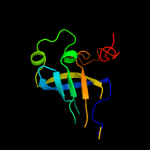
1 c1e3hA_
100.0
45
PDB header: polyribonucleotide transferaseChain: A: PDB Molecule: guanosine pentaphosphate synthetase;PDBTitle: semet derivative of streptomyces antibioticus pnpase/gpsi2 enzyme
2 c3cdiA_
100.0
100
PDB header: transferaseChain: A: PDB Molecule: polynucleotide phosphorylase;PDBTitle: crystal structure of e. coli pnpase
3 c3cdjA_
100.0
100
PDB header: transferaseChain: A: PDB Molecule: polynucleotide phosphorylase;PDBTitle: crystal structure of the e. coli kh/s1 domain truncated2 pnpase
4 c2po2A_
100.0
28
PDB header: hydrolase/hydrolaseChain: A: PDB Molecule: probable exosome complex exonuclease 1;PDBTitle: crystal structure of the p. abyssi exosome rnase ph ring2 complexed with cdp
5 c2wnrB_
100.0
31
PDB header: hydrolaseChain: B: PDB Molecule: probable exosome complex exonuclease 1;PDBTitle: the structure of methanothermobacter thermautotrophicus2 exosome core assembly
6 c2nn6B_
100.0
22
PDB header: hydrolase/transferaseChain: B: PDB Molecule: exosome complex exonuclease rrp41;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
7 c2ba1D_
100.0
26
PDB header: rna binding proteinChain: D: PDB Molecule: archaeal exosome complex exonuclease rrp41;PDBTitle: archaeal exosome core
8 c2c37L_
100.0
27
PDB header: hydrolaseChain: L: PDB Molecule: probable exosome complex exonuclease 1;PDBTitle: rnase ph core of the archaeal exosome in complex with u82 rna
9 c3dd6A_
100.0
25
PDB header: transferaseChain: A: PDB Molecule: ribonuclease ph;PDBTitle: crystal structure of rph, an exoribonuclease from bacillus2 anthracis at 1.7 a resolution
10 c2wp8B_
100.0
20
PDB header: hydrolaseChain: B: PDB Molecule: exosome complex component ski6;PDBTitle: yeast rrp44 nuclease
11 c1udsA_
100.0
24
PDB header: transferaseChain: A: PDB Molecule: ribonuclease ph;PDBTitle: crystal structure of the trna processing enzyme rnase ph r126a mutant2 from aquifex aeolicus
12 c2pnzB_
100.0
20
PDB header: hydrolase/hydrolaseChain: B: PDB Molecule: probable exosome complex exonuclease 2;PDBTitle: crystal structure of the p. abyssi exosome rnase ph ring2 complexed with udp and gmp
13 c3b4tC_
100.0
24
PDB header: transferaseChain: C: PDB Molecule: ribonuclease ph;PDBTitle: crystal structure of mycobacterium tuberculosis rnase ph, the2 mycobacterium tuberculosis structural genomics consortium target3 rv1340
14 c1r6mA_
100.0
25
PDB header: transferaseChain: A: PDB Molecule: ribonuclease ph;PDBTitle: crystal structure of the trna processing enzyme rnase ph from2 pseudomonas aeruginosa in complex with phosphate
15 d1e3ha2
100.0
49
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like16 c2nn6C_
100.0
18
PDB header: hydrolase/transferaseChain: C: PDB Molecule: exosome complex exonuclease rrp43;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
17 c2wnrC_
100.0
18
PDB header: hydrolaseChain: C: PDB Molecule: probable exosome complex exonuclease 2;PDBTitle: the structure of methanothermobacter thermautotrophicus2 exosome core assembly
18 c3hkmB_
100.0
23
PDB header: hydrolaseChain: B: PDB Molecule: os03g0854200 protein;PDBTitle: crystal structure of rice(oryza sativa) rrp46
19 c2ba0I_
100.0
22
PDB header: rna binding proteinChain: I: PDB Molecule: archeal exosome rna binding protein rrp42;PDBTitle: archaeal exosome core
20 c2nn6E_
100.0
17
PDB header: hydrolase/transferaseChain: E: PDB Molecule: exosome complex exonuclease rrp42;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
21 c2br2G_
not modelled
100.0
20
PDB header: hydrolaseChain: G: PDB Molecule: exosome complex exonuclease 2;PDBTitle: rnase ph core of the archaeal exosome
22 c2nn6D_
not modelled
100.0
24
PDB header: hydrolase/transferaseChain: D: PDB Molecule: exosome complex exonuclease rrp46;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
23 c2nn6F_
not modelled
100.0
21
PDB header: hydrolase/transferaseChain: F: PDB Molecule: exosome component 6;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
24 d1e3ha6
not modelled
100.0
51
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like25 d1e3ha3
not modelled
100.0
51
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like26 c2wp8A_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: exosome complex component rrp45;PDBTitle: yeast rrp44 nuclease
27 d2je6b1
not modelled
100.0
30
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like28 d2ba0d1
not modelled
100.0
29
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like29 d2nn6b1
not modelled
100.0
23
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like30 d1r6la1
not modelled
100.0
25
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like31 d1udsa1
not modelled
100.0
22
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like32 d2ba0g1
not modelled
100.0
25
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like33 d2je6a1
not modelled
99.9
19
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like34 d2nn6e1
not modelled
99.9
20
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like35 c3krnB_
not modelled
99.9
18
PDB header: hydrolaseChain: B: PDB Molecule: protein c14a4.5, confirmed by transcript evidence;PDBTitle: crystal structure of c. elegans cell-death-related nuclease 5(crn-5)
36 d2nn6a1
not modelled
99.9
17
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like37 d2nn6c1
not modelled
99.9
17
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like38 d1oysa1
not modelled
99.9
30
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like39 d2nn6f1
not modelled
99.9
25
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like40 d1e3ha5
not modelled
99.9
33
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like41 d2nn6d1
not modelled
99.9
31
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like42 d1udsa2
not modelled
99.8
19
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like43 d1r6la2
not modelled
99.7
19
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like44 d1whua_
not modelled
99.7
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3Family: Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 345 d1oysa2
not modelled
99.7
23
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like46 c2cqoA_
99.6
27
PDB header: ribosomeChain: A: PDB Molecule: nucleolar protein of 40 kda;PDBTitle: solution structure of the s1 rna binding domain of human2 hypothetical protein flj11067
47 c1q46A_
99.6
24
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
48 c1yz6A_
99.6
42
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 alphaPDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
49 d1e3ha1
not modelled
99.5
35
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3Family: Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 350 c1q8kA_
not modelled
99.5
29
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2PDBTitle: solution structure of alpha subunit of human eif2
51 d2ba0d2
not modelled
99.5
20
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like52 c2k4kA_
99.5
40
PDB header: rna binding proteinChain: A: PDB Molecule: general stress protein 13;PDBTitle: solution structure of gsp13 from bacillus subtilis
53 d1q46a2
not modelled
99.5
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like54 c2eqsA_
99.5
33
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dhx8;PDBTitle: solution structure of the s1 rna binding domain of human2 atp-dependent rna helicase dhx8
55 d2je6b2
not modelled
99.5
16
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like56 d2br2b2
not modelled
99.5
15
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like57 c2khjA_
99.4
35
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
58 d2ba0a1
not modelled
99.4
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like59 d1kl9a2
not modelled
99.4
37
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like60 c2oceA_
not modelled
99.4
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa5201;PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
61 c2khiA_
not modelled
99.4
32
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
62 d1go3e1
not modelled
99.4
29
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like63 d2nn6b2
not modelled
99.4
15
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like64 d1sroa_
not modelled
99.4
100
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like65 c2ahoB_
not modelled
99.4
30
PDB header: translationChain: B: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: structure of the archaeal initiation factor eif2 alpha-2 gamma heterodimer from sulfolobus solfataricus complexed3 with gdpnp
66 c2k52A_
not modelled
99.3
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mj1198;PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
67 d2z0sa1
not modelled
99.3
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like68 c1kl9A_
not modelled
99.3
33
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2 subunit 1;PDBTitle: crystal structure of the n-terminal segment of human eukaryotic2 initiation factor 2alpha
69 d3bzka4
not modelled
99.3
41
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like70 d2nn6h1
not modelled
99.3
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like71 d2je6i1
not modelled
99.3
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like72 c3psiA_
not modelled
99.3
15
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(239-1451)
73 c1hh2P_
99.2
18
PDB header: transcription regulationChain: P: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima
74 d2z0sa2
99.2
25
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)75 d2ahob2
not modelled
99.2
31
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like76 d2je6a2
not modelled
99.2
16
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like77 d2nn6d2
not modelled
99.2
21
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like78 d1smxa_
not modelled
99.1
31
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like79 d1wi5a_
not modelled
99.1
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like80 c3go5A_
not modelled
99.0
19
PDB header: gene regulationChain: A: PDB Molecule: multidomain protein with s1 rna-binding domains;PDBTitle: crystal structure of a multidomain protein with nucleic acid binding2 domains (sp_0946) from streptococcus pneumoniae tigr4 at 1.40 a3 resolution
81 c2z0sA_
not modelled
99.0
18
PDB header: rna binding proteinChain: A: PDB Molecule: probable exosome complex rna-binding protein 1;PDBTitle: crystal structure of putative exosome complex rna-binding2 protein
82 d1y14b1
not modelled
99.0
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like83 c2ba0A_
not modelled
99.0
23
PDB header: rna binding proteinChain: A: PDB Molecule: archeal exosome rna binding protein rrp4;PDBTitle: archaeal exosome core
84 c2bh8B_
not modelled
99.0
36
PDB header: transcriptionChain: B: PDB Molecule: 1b11;PDBTitle: combinatorial protein 1b11
85 d2ba0a3
not modelled
98.9
31
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)86 d2nn6i1
not modelled
98.9
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like87 d2nn6f2
not modelled
98.9
18
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like88 c2je6I_
not modelled
98.8
23
PDB header: hydrolaseChain: I: PDB Molecule: exosome complex rna-binding protein 1;PDBTitle: structure of a 9-subunit archaeal exosome
89 c1go3E_
not modelled
98.7
29
PDB header: transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: structure of an archeal homolog of the eukaryotic rna2 polymerase ii rpb4/rpb7 complex
90 d1hh2p1
not modelled
98.7
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like91 c2ba1B_
not modelled
98.7
23
PDB header: rna binding proteinChain: B: PDB Molecule: archaeal exosome rna binding protein csl4;PDBTitle: archaeal exosome core
92 d2ctka1
not modelled
98.6
20
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)93 d1tuaa1
not modelled
98.6
25
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)94 c2nn6I_
not modelled
98.6
22
PDB header: hydrolase/transferaseChain: I: PDB Molecule: 3'-5' exoribonuclease csl4 homolog;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
95 d2ba0g2
not modelled
98.6
14
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like96 c1l2fA_
not modelled
98.5
18
PDB header: transcriptionChain: A: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima: a2 structure-based role of the n-terminal domain
97 c2pmzE_
not modelled
98.5
41
PDB header: translation, transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
98 d2c35b1
not modelled
98.5
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like99 c2c35F_
not modelled
98.5
19
PDB header: polymeraseChain: F: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: subunits rpb4 and rpb7 of human rna polymerase ii
100 c2e3uA_
not modelled
98.5
22
PDB header: rna binding proteinChain: A: PDB Molecule: hypothetical protein ph1566;PDBTitle: crystal structure analysis of dim2p from pyrococcus horikoshii ot3
101 d1we8a_
not modelled
98.5
24
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)102 c2yqrA_
not modelled
98.4
20
PDB header: rna binding proteinChain: A: PDB Molecule: kiaa0907 protein;PDBTitle: solution structure of the kh domain in kiaa0907 protein
103 d1j4wa1
not modelled
98.4
23
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)104 d1x4ma1
not modelled
98.4
33
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)105 c3h0gS_
not modelled
98.4
21
PDB header: transcriptionChain: S: PDB Molecule: dna-directed rna polymerase ii subunit rpb7;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
106 c3n89B_
not modelled
98.4
13
PDB header: cell cycleChain: B: PDB Molecule: defective in germ line development protein 3, isoform a;PDBTitle: kh domains
107 c2hh2A_
not modelled
98.4
32
PDB header: rna binding proteinChain: A: PDB Molecule: kh-type splicing regulatory protein;PDBTitle: solution structure of the fourth kh domain of ksrp
108 c2b8kG_
not modelled
98.4
16
PDB header: transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: 12-subunit rna polymerase ii
109 d1khma_
not modelled
98.4
22
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)110 d2ctfa1
not modelled
98.4
17
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)111 d1dtja_
not modelled
98.4
24
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)112 d1zzka1
not modelled
98.3
22
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)113 d1ec6a_
not modelled
98.3
25
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)114 c2ckzB_
not modelled
98.3
12
PDB header: transferaseChain: B: PDB Molecule: dna-directed rna polymerase iii 25 kdPDBTitle: x-ray structure of rna polymerase iii subcomplex c17-c25.
115 d1x4na1
not modelled
98.3
23
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)116 d1viga_
not modelled
98.3
22
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)117 c2dgrA_
not modelled
98.3
17
PDB header: rna binding proteinChain: A: PDB Molecule: ring finger and kh domain-containing protein 1;PDBTitle: solution structure of the second kh domain in ring finger2 and kh domain containing protein 1
118 d1wvna1
not modelled
98.3
26
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)119 d2axya1
not modelled
98.3
19
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)120 d2nn6a2
not modelled
98.3
13
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like