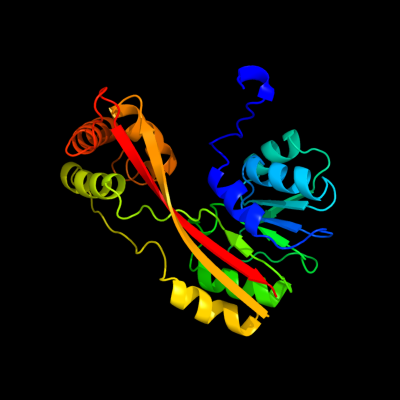
| 1 | c2p35A_
|
|
 |
100.0 |
44 |
PDB header:transferase
Chain: A: PDB Molecule:trans-aconitate 2-methyltransferase;
PDBTitle: crystal structure of trans-aconitate methyltransferase from2 agrobacterium tumefaciens
|
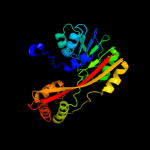
| 2 | c3ccfB_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:cyclopropane-fatty-acyl-phospholipid synthase;
PDBTitle: crystal structure of putative methyltransferase (yp_321342.1) from2 anabaena variabilis atcc 29413 at 1.90 a resolution
|
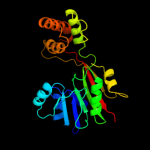
| 3 | c3g5tA_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:trans-aconitate 3-methyltransferase;
PDBTitle: crystal structure of trans-aconitate 3-methyltransferase2 from yeast
|
| 4 | c2yr0A_
|
|
 |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein ttha0223;
PDBTitle: crystal structure of hypothetical methyltransferase ttha0223 from2 thermus thermophilus hb8
|
| 5 | c1vl5B_
|
|
 |
99.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:unknown conserved protein bh2331;
PDBTitle: crystal structure of a putative methyltransferase (bh2331) from2 bacillus halodurans c-125 at 1.95 a resolution
|
| 6 | c3mggB_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: crystal structure of methyl transferase from methanosarcina2 mazei
|
| 7 | d1vl5a_
|
|
 |
99.9 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:UbiE/COQ5-like |
| 8 | c3bkxB_
|
|
 |
99.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of cyclopropane-fatty-acyl-phospholipid synthase-2 like protein (yp_807781.1) from lactobacillus casei atcc 334 at 1.853 a resolution
|
| 9 | d2o57a1
|
|
 |
99.9 |
15 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 10 | d1jqea_
|
|
 |
99.9 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Histamine methyltransferase |
| 11 | c3busB_
|
|
 |
99.9 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: crystal structure of rebm
|
| 12 | d1xxla_
|
|
 |
99.9 |
11 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:UbiE/COQ5-like |
| 13 | c2fk8A_
|
|
 |
99.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:methoxy mycolic acid synthase 4;
PDBTitle: crystal structure of hma (mmaa4) from mycobacterium tuberculosis2 complexed with s-adenosylmethionine
|
| 14 | d1nkva_
|
|
 |
99.9 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Hypothetical Protein YjhP |
| 15 | d2fk8a1
|
|
 |
99.9 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 16 | c3ujcA_
|
|
 |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoethanolamine n-methyltransferase;
PDBTitle: phosphoethanolamine methyltransferase mutant (h132a) from plasmodium2 falciparum in complex with phosphocholine
|
| 17 | c3cc8A_
|
|
 |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase;
PDBTitle: crystal structure of a putative methyltransferase (bce_1332) from2 bacillus cereus atcc 10987 at 1.64 a resolution
|
| 18 | d1kpga_
|
|
 |
99.8 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 19 | c3e7pA_
|
|
 |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase;
PDBTitle: crystal structure of of putative methyltransferase from bacteroides2 vulgatus atcc 8482
|
| 20 | d1kpia_
|
|
 |
99.8 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 21 | c3egeA_ |
|
not modelled |
99.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase from antibiotic biosynthesis
PDBTitle: crystal structure of putative methyltransferase from antibiotic2 biosynthesis pathway (yp_324569.1) from anabaena variabilis atcc3 29413 at 2.40 a resolution
|
| 22 | d1l1ea_ |
|
not modelled |
99.8 |
11 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 23 | c3f4kA_ |
|
not modelled |
99.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase;
PDBTitle: crystal structure of a probable methyltransferase from2 bacteroides thetaiotaomicron. northeast structural3 genomics target btr309.
|
| 24 | d1tpya_ |
|
not modelled |
99.8 |
11 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 25 | c3dtnA_ |
|
not modelled |
99.8 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative methyltransferase mm_2633;
PDBTitle: crystal structure of putative methyltransferase-mm_26332 from methanosarcina mazei .
|
| 26 | c3e23A_ |
|
not modelled |
99.8 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein rpa2492;
PDBTitle: crystal structure of the rpa2492 protein in complex with2 sam from rhodopseudomonas palustris, northeast structural3 genomics consortium target rpr299
|
| 27 | c3dlcA_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative s-adenosyl-l-methionine-dependent
PDBTitle: crystal structure of a putative s-adenosyl-l-methionine-dependent2 methyltransferase (mmp1179) from methanococcus maripaludis at 1.15 a3 resolution
|
| 28 | d2gh1a1 |
|
not modelled |
99.8 |
22 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:BC2162-like |
| 29 | c2gs9A_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein tt1324;
PDBTitle: crystal structure of tt1324 from thermus thermophilis hb8
|
| 30 | c3l8dA_ |
|
not modelled |
99.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: crystal structure of methyltransferase from bacillus2 thuringiensis
|
| 31 | d2p7ia1 |
|
not modelled |
99.8 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:UbiE/COQ5-like |
| 32 | d1vlma_ |
|
not modelled |
99.8 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:UbiE/COQ5-like |
| 33 | c3e8sA_ |
|
not modelled |
99.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative sam dependent methyltransferase;
PDBTitle: crystal structure of putative sam dependent methyltransferase in2 complex with sah (np_744700.1) from pseudomonas putida kt2440 at 2.103 a resolution
|
| 34 | c3dliB_ |
|
not modelled |
99.7 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: crystal structure of a sam dependent methyltransferase from2 archaeoglobus fulgidus
|
| 35 | c3ou7A_ |
|
not modelled |
99.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: dhpi-sam-hep complex
|
| 36 | c2p7iB_ |
|
not modelled |
99.7 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a sam dependent methyl-transferase type 12 family2 protein (eca1738) from pectobacterium atrosepticum scri1043 at 1.74 a3 resolution
|
| 37 | d1p91a_ |
|
not modelled |
99.7 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:rRNA methyltransferase RlmA |
| 38 | d1im8a_ |
|
not modelled |
99.7 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Hypothetical protein HI0319 (YecO) |
| 39 | c3mq2A_ |
|
not modelled |
99.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:16s rrna methyltransferase;
PDBTitle: crystal structure of 16s rrna methyltranferase kamb
|
| 40 | d2avna1 |
|
not modelled |
99.7 |
22 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:UbiE/COQ5-like |
| 41 | c3pfhD_ |
|
not modelled |
99.7 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:n-methyltransferase;
PDBTitle: x-ray crystal structure the n,n-dimethyltransferase tylm1 from2 streptomyces fradiae in complex with sah and dtdp-quip3n
|
| 42 | c3ocjA_ |
|
not modelled |
99.7 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: the crystal structure of a possilbe exported protein from bordetella2 parapertussis
|
| 43 | c3dh0B_ |
|
not modelled |
99.7 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:sam dependent methyltransferase;
PDBTitle: crystal structure of a sam dependent methyltransferase from2 aquifex aeolicus
|
| 44 | c3ofkA_ |
|
not modelled |
99.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:nodulation protein s;
PDBTitle: crystal structure of n-methyltransferase nods from bradyrhizobium2 japonicum wm9 in complex with s-adenosyl-l-homocysteine (sah)
|
| 45 | d1xtpa_ |
|
not modelled |
99.7 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:AD-003 protein-like |
| 46 | c3hnrA_ |
|
not modelled |
99.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:probable methyltransferase bt9727_4108;
PDBTitle: crystal structure of a probable methyltransferase2 bt9727_4108 from bacillus thuringiensis subsp. northeast3 structural genomics consortium target id bur219
|
| 47 | c3h2bB_ |
|
not modelled |
99.7 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of the sam-dependent methyltransferase2 cg3271 from corynebacterium glutamicum in complex with s-3 adenosyl-l-homocysteine and pyrophosphate. northeast4 structural genomics consortium target cgr113a
|
| 48 | d1kyza2 |
|
not modelled |
99.7 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 49 | d1wzna1 |
|
not modelled |
99.7 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:CAC2371-like |
| 50 | c3ndjA_ |
|
not modelled |
99.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: x-ray structure of a c-3'-methyltransferase in complex with s-2 adenosyl-l-homocysteine and sugar product
|
| 51 | c3bgvC_ |
|
not modelled |
99.7 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:mrna cap guanine-n7 methyltransferase;
PDBTitle: crystal structure of mrna cap guanine-n7 methyltransferase2 in complex with sah
|
| 52 | c3g5lA_ |
|
not modelled |
99.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative s-adenosylmethionine dependent
PDBTitle: crystal structure of putative s-adenosylmethionine2 dependent methyltransferase from listeria monocytogenes
|
| 53 | c3bkwB_ |
|
not modelled |
99.7 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine dependent methyltransferase;
PDBTitle: crystal structure of s-adenosylmethionine dependent methyltransferase2 (np_104914.1) from mesorhizobium loti at 1.60 a resolution
|
| 54 | c3lstB_ |
|
not modelled |
99.7 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:calo1 methyltransferase;
PDBTitle: crystal structure of calo1, methyltransferase in calicheamicin2 biosynthesis, sah bound form
|
| 55 | d1fp1d2 |
|
not modelled |
99.7 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 56 | c3lccA_ |
|
not modelled |
99.6 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyl chloride transferase;
PDBTitle: structure of a sam-dependent halide methyltransferase from arabidopsis2 thaliana
|
| 57 | c3bxoA_ |
|
not modelled |
99.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:n,n-dimethyltransferase;
PDBTitle: crystal structure of streptomyces venezuelae desvi
|
| 58 | d2ex4a1 |
|
not modelled |
99.6 |
16 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:AD-003 protein-like |
| 59 | d1yzha1 |
|
not modelled |
99.6 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TrmB-like |
| 60 | d1y8ca_ |
|
not modelled |
99.6 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:CAC2371-like |
| 61 | c3mczB_ |
|
not modelled |
99.6 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:o-methyltransferase;
PDBTitle: the structure of an o-methyltransferase family protein from2 burkholderia thailandensis.
|
| 62 | c2r3sA_ |
|
not modelled |
99.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative o-methyltransferase (npun_r0239) from2 nostoc punctiforme pcc 73102 at 2.15 a resolution
|
| 63 | d1ri5a_ |
|
not modelled |
99.6 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:mRNA cap (Guanine N-7) methyltransferase |
| 64 | c1z3cA_ |
|
not modelled |
99.6 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:mrna capping enzyme;
PDBTitle: encephalitozooan cuniculi mrna cap (guanine-n7)2 methyltransferasein complexed with azoadomet
|
| 65 | c3opnA_ |
|
not modelled |
99.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative hemolysin;
PDBTitle: the crystal structure of a putative hemolysin from lactococcus lactis
|
| 66 | d1qzza2 |
|
not modelled |
99.6 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 67 | c3dp7B_ |
|
not modelled |
99.6 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of sam-dependent methyltransferase from bacteroides2 vulgatus atcc 8482
|
| 68 | c3g2qA_ |
|
not modelled |
99.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:pcza361.24;
PDBTitle: crystal structure of the glycopeptide n-methyltransferase mtfa2 complexed with sinefungin
|
| 69 | d1ve3a1 |
|
not modelled |
99.6 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:CAC2371-like |
| 70 | d1tw3a2 |
|
not modelled |
99.6 |
16 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 71 | c3sm3A_ |
|
not modelled |
99.6 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:sam-dependent methyltransferases;
PDBTitle: crystal structure of sam-dependent methyltransferases q8puk2_metma2 from methanosarcina mazei. northeast structural genomics consortium3 target mar262.
|
| 72 | d2a14a1 |
|
not modelled |
99.6 |
15 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Arylamine N-methyltransferase |
| 73 | d1xcla_ |
|
not modelled |
99.6 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Guanidinoacetate methyltransferase |
| 74 | c3gwzB_ |
|
not modelled |
99.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:mmcr;
PDBTitle: structure of the mitomycin 7-o-methyltransferase mmcr
|
| 75 | d1m6ya2 |
|
not modelled |
99.6 |
22 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:MraW-like putative methyltransferases |
| 76 | c3merA_ |
|
not modelled |
99.6 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:slr1183 protein;
PDBTitle: crystal structure of the methyltransferase slr1183 from2 synechocystis sp. pcc 6803, northeast structural genomics3 consortium target sgr145
|
| 77 | c3p9kD_ |
|
not modelled |
99.6 |
15 |
PDB header:transferase
Chain: D: PDB Molecule:caffeic acid o-methyltransferase;
PDBTitle: crystal structure of perennial ryegrass lpomt1 complexed with s-2 adenosyl-l-homocysteine and coniferaldehyde
|
| 78 | d1zx0a1 |
|
not modelled |
99.6 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Guanidinoacetate methyltransferase |
| 79 | c1kyzC_ |
|
not modelled |
99.6 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:caffeic acid 3-o-methyltransferase;
PDBTitle: crystal structure analysis of caffeic acid/5-hydroxyferulic2 acid 3/5-o-methyltransferase ferulic acid complex
|
| 80 | c3i53A_ |
|
not modelled |
99.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:o-methyltransferase;
PDBTitle: crystal structure of an o-methyltransferase (ncsb1) from2 neocarzinostatin biosynthesis in complex with s-adenosyl-l-3 homocysteine (sah)
|
| 81 | c3ggdA_ |
|
not modelled |
99.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of sam-dependent methyltransferase (yp_325210.1)2 from anabaena variabilis atcc 29413 at 2.11 a resolution
|
| 82 | d1xvaa_ |
|
not modelled |
99.6 |
22 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Glycine N-methyltransferase |
| 83 | c2iipD_ |
|
not modelled |
99.6 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinamide n-methyltransferase;
PDBTitle: human nicotinamide n-methyltransferase
|
| 84 | c1x1aA_ |
|
not modelled |
99.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:crtf-related protein;
PDBTitle: crystal structure of bchu complexed with s-adenosyl-l-methionine
|
| 85 | d1d2ha_ |
|
not modelled |
99.5 |
22 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Glycine N-methyltransferase |
| 86 | d2fcaa1 |
|
not modelled |
99.5 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TrmB-like |
| 87 | c2zfuA_ |
|
not modelled |
99.5 |
15 |
PDB header:nuclear protein
Chain: A: PDB Molecule:cerebral protein 1;
PDBTitle: structure of the methyltransferase-like domain of nucleomethylin
|
| 88 | c2ip2B_ |
|
not modelled |
99.5 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:probable phenazine-specific methyltransferase;
PDBTitle: structure of the pyocyanin biosynthetic protein phzm
|
| 89 | c3g07C_ |
|
not modelled |
99.5 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:7sk snrna methylphosphate capping enzyme;
PDBTitle: methyltransferase domain of human bicoid-interacting protein2 3 homolog (drosophila)
|
| 90 | c3cggB_ |
|
not modelled |
99.5 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of tehb-like sam-dependent methyltransferase2 (np_600671.1) from corynebacterium glutamicum atcc 13032 kitasato at3 2.00 a resolution
|
| 91 | c3jwgA_ |
|
not modelled |
99.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase type 12;
PDBTitle: crystal structure analysis of the methyltransferase domain2 of bacterial-cthen1-c
|
| 92 | d2i6ga1 |
|
not modelled |
99.5 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TehB-like |
| 93 | c3lecA_ |
|
not modelled |
99.5 |
11 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:nadb-rossmann superfamily protein;
PDBTitle: the crystal structure of a protein in the nadb-rossmann2 superfamily from streptococcus agalactiae to 1.8a
|
| 94 | c3i9fB_ |
|
not modelled |
99.5 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:putative type 11 methyltransferase;
PDBTitle: crystal structure of a putative type 11 methyltransferase2 from sulfolobus solfataricus
|
| 95 | d1g8sa_ |
|
not modelled |
99.5 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Fibrillarin homologue |
| 96 | c3dxyA_ |
|
not modelled |
99.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanine-n(7)-)-methyltransferase;
PDBTitle: crystal structure of ectrmb in complex with sam
|
| 97 | c3d2lC_ |
|
not modelled |
99.5 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of sam-dependent methyltransferase (zp_00538691.1)2 from exiguobacterium sp. 255-15 at 1.90 a resolution
|
| 98 | d1pjza_ |
|
not modelled |
99.5 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Thiopurine S-methyltransferase |
| 99 | d1dusa_ |
|
not modelled |
99.5 |
16 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Hypothetical protein MJ0882 |
| 100 | d1fp2a2 |
|
not modelled |
99.5 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 101 | d1yb2a1 |
|
not modelled |
99.5 |
20 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:tRNA(1-methyladenosine) methyltransferase-like |
| 102 | c1yb2A_ |
|
not modelled |
99.5 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ta0852;
PDBTitle: structure of a putative methyltransferase from thermoplasma2 acidophilum.
|
| 103 | d1r74a_ |
|
not modelled |
99.5 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Glycine N-methyltransferase |
| 104 | c3p2kA_ |
|
not modelled |
99.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:16s rrna methylase;
PDBTitle: structure of an antibiotic related methyltransferase
|
| 105 | c3mtiA_ |
|
not modelled |
99.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:rrna methylase;
PDBTitle: the crystal structure of a rrna methylase from streptococcus2 thermophilus to 1.95a
|
| 106 | d1p1ca_ |
|
not modelled |
99.5 |
14 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Guanidinoacetate methyltransferase |
| 107 | c1tw3A_ |
|
not modelled |
99.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:carminomycin 4-o-methyltransferase;
PDBTitle: crystal structure of carminomycin-4-o-methyltransferase2 (dnrk) in complex with s-adenosyl-l-homocystein (sah) and3 4-methoxy-e-rhodomycin t (m-et)
|
| 108 | c1zgaA_ |
|
not modelled |
99.5 |
16 |
PDB header:plant protein, transferase
Chain: A: PDB Molecule:isoflavanone 4'-o-methyltransferase';
PDBTitle: crystal structure of isoflavanone 4'-o-methyltransferase complexed2 with (+)-6a-hydroxymaackiain
|
| 109 | c3m70A_ |
|
not modelled |
99.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein tehb homolog;
PDBTitle: crystal structure of tehb from haemophilus influenzae
|
| 110 | d1o54a_ |
|
not modelled |
99.5 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:tRNA(1-methyladenosine) methyltransferase-like |
| 111 | c3mb5A_ |
|
not modelled |
99.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of p. abyssi trna m1a58 methyltransferase in complex2 with s-adenosyl-l-methionine
|
| 112 | c3gnlB_ |
|
not modelled |
99.5 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein, duf633, lmof2365_1472;
PDBTitle: structure of uncharacterized protein (lmof2365_1472) from2 listeria monocytogenes serotype 4b
|
| 113 | c1xduA_ |
|
not modelled |
99.5 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:protein rdmb;
PDBTitle: crystal structure of aclacinomycin-10-hydroxylase (rdmb) in complex2 with sinefungin (sfg)
|
| 114 | d1wg8a2 |
|
not modelled |
99.5 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:MraW-like putative methyltransferases |
| 115 | c3eeyJ_ |
|
not modelled |
99.5 |
17 |
PDB header:transferase
Chain: J: PDB Molecule:putative rrna methylase;
PDBTitle: crystal structure of putative rrna-methylase from clostridium2 thermocellum
|
| 116 | c3jwhA_ |
|
not modelled |
99.4 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:hen1;
PDBTitle: crystal structure analysis of the methyltransferase domain2 of bacterial-avhen1-c
|
| 117 | c2yvlB_ |
|
not modelled |
99.4 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of trna (m1a58) methyltransferase trmi from aquifex2 aeolicus
|
| 118 | d2nxca1 |
|
not modelled |
99.4 |
25 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Ribosomal protein L11 methyltransferase PrmA |
| 119 | c1fpqA_ |
|
not modelled |
99.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:isoliquiritigenin 2'-o-methyltransferase;
PDBTitle: crystal structure analysis of selenomethionine substituted chalcone o-2 methyltransferase
|
| 120 | c3e05B_ |
|
not modelled |
99.4 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-6y c5,15-methyltransferase (decarboxylating);
PDBTitle: crystal structure of precorrin-6y c5,15-methyltransferase from2 geobacter metallireducens gs-15
|