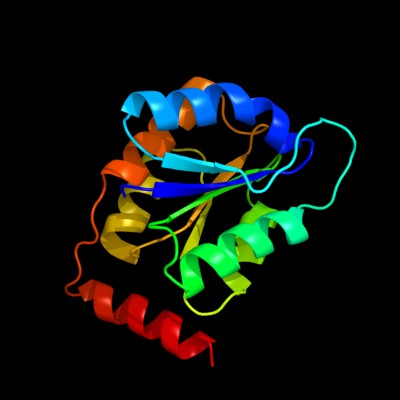
| 1 | d1nn4a_
|
|
 |
100.0 |
100 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
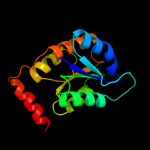
| 2 | c3he8A_
|
|
 |
100.0 |
57 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
|
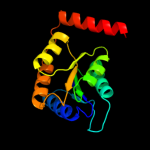
| 3 | c3m1pA_
|
|
 |
100.0 |
40 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
|
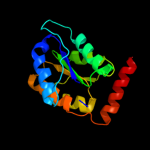
| 4 | c3k7pA_
|
|
 |
100.0 |
40 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
|
| 5 | d2vvpa1
|
|
 |
100.0 |
32 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
| 6 | d1o1xa_
|
|
 |
100.0 |
46 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
| 7 | c3qd5B_
|
|
 |
100.0 |
39 |
PDB header:isomerase
Chain: B: PDB Molecule:putative ribose-5-phosphate isomerase;
PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
|
| 8 | c3s5pA_
|
|
 |
100.0 |
45 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
|
| 9 | c2ppwA_
|
|
 |
100.0 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
|
| 10 | c3onoA_
|
|
 |
100.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
|
| 11 | c3c5yD_
|
|
 |
100.0 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
|
| 12 | c2pjkA_
|
|
 |
93.2 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:178aa long hypothetical molybdenum cofactor
PDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
|
| 13 | d1mkza_
|
|
 |
91.5 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 14 | d1y5ea1
|
|
 |
91.0 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 15 | d1p5dx1
|
|
 |
90.0 |
27 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 16 | c3pdkB_
|
|
 |
89.0 |
29 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: crystal structure of phosphoglucosamine mutase from b. anthracis
|
| 17 | c3i3wB_
|
|
 |
89.0 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: structure of a phosphoglucosamine mutase from francisella tularensis
|
| 18 | c2is8A_
|
|
 |
88.5 |
20 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
| 19 | c1wqaB_
|
|
 |
88.3 |
31 |
PDB header:isomerase
Chain: B: PDB Molecule:phospho-sugar mutase;
PDBTitle: crystal structure of pyrococcus horikoshii2 phosphomannomutase/phosphoglucomutase complexed with mg2+
|
| 20 | c2f7lA_
|
|
 |
87.8 |
34 |
PDB header:isomerase
Chain: A: PDB Molecule:455aa long hypothetical phospho-sugar mutase;
PDBTitle: crystal structure of sulfolobus tokodaii2 phosphomannomutase/phosphoglucomutase
|
| 21 | c3c04A_ |
|
not modelled |
82.0 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphomannomutase/phosphoglucomutase;
PDBTitle: structure of the p368g mutant of pmm/pgm from p. aeruginosa
|
| 22 | c2xecD_ |
|
not modelled |
81.4 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 23 | c1tvmA_ |
|
not modelled |
80.8 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, galactitol-specific iib component;
PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
|
| 24 | c1tuoA_ |
|
not modelled |
80.3 |
22 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative phosphomannomutase;
PDBTitle: crystal structure of putative phosphomannomutase from2 thermus thermophilus hb8
|
| 25 | d1jlja_ |
|
not modelled |
79.6 |
10 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 26 | c3rfqC_ |
|
not modelled |
78.3 |
29 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
| 27 | d1uuya_ |
|
not modelled |
77.7 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 28 | d2g2ca1 |
|
not modelled |
74.2 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 29 | c1vkrA_ |
|
not modelled |
70.8 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific pts system enzyme iiabc components;
PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
|
| 30 | d1vkra_ |
|
not modelled |
70.8 |
32 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 31 | c3czcA_ |
|
not modelled |
70.2 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:rmpb;
PDBTitle: the crystal structure of a putative pts iib(ptxb) from2 streptococcus mutans
|
| 32 | d2gaxa1 |
|
not modelled |
68.6 |
10 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
| 33 | c3kbqA_ |
|
not modelled |
67.7 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ta0487;
PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum
|
| 34 | d2ftsa3 |
|
not modelled |
67.1 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 35 | d2f7wa1 |
|
not modelled |
66.7 |
9 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 36 | c2yxbA_ |
|
not modelled |
65.7 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 37 | c3gpiA_ |
|
not modelled |
61.6 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: structure of putative nad-dependent epimerase/dehydratase2 from methylobacillus flagellatus
|
| 38 | c3bq9A_ |
|
not modelled |
61.2 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
|
| 39 | c2dgdD_ |
|
not modelled |
60.5 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:223aa long hypothetical arylmalonate decarboxylase;
PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
|
| 40 | d3bula2 |
|
not modelled |
60.1 |
22 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 41 | d1v7ra_ |
|
not modelled |
59.1 |
18 |
Fold:Anticodon-binding domain-like
Superfamily:ITPase-like
Family:ITPase (Ham1) |
| 42 | d1uz5a3 |
|
not modelled |
58.4 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 43 | c1k98A_ |
|
not modelled |
54.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
| 44 | d2nqra3 |
|
not modelled |
46.4 |
29 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 45 | d1di6a_ |
|
not modelled |
46.3 |
9 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 46 | c3ezxA_ |
|
not modelled |
45.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
| 47 | d1fmfa_ |
|
not modelled |
44.5 |
30 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 48 | c2nqqA_ |
|
not modelled |
44.3 |
29 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
| 49 | c1c4gB_ |
|
not modelled |
41.5 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:protein (alpha-d-glucose 1-phosphate
PDBTitle: phosphoglucomutase vanadate based transition state analog2 complex
|
| 50 | c2g4rB_ |
|
not modelled |
41.4 |
26 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis mog protein;
PDBTitle: anomalous substructure of moga
|
| 51 | c1kfiA_ |
|
not modelled |
40.9 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase 1;
PDBTitle: crystal structure of the exocytosis-sensitive2 phosphoprotein, pp63/parafusin (phosphoglucomutase) from3 paramecium
|
| 52 | d1ws6a1 |
|
not modelled |
40.4 |
15 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
| 53 | c1bmtB_ |
|
not modelled |
38.1 |
22 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
| 54 | c2r60A_ |
|
not modelled |
37.8 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase, group 1;
PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
|
| 55 | c2fuvB_ |
|
not modelled |
37.0 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucomutase;
PDBTitle: phosphoglucomutase from salmonella typhimurium.
|
| 56 | c3dz1A_ |
|
not modelled |
35.8 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 57 | d7reqa2 |
|
not modelled |
35.3 |
17 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 58 | d1xova2 |
|
not modelled |
34.9 |
14 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 59 | c3ir1F_ |
|
not modelled |
34.1 |
26 |
PDB header:protein binding
Chain: F: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
|
| 60 | c3tngA_ |
|
not modelled |
33.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:lmo1369 protein;
PDBTitle: the crystal structure of a possible phosphate acetyl/butaryl2 transferase from listeria monocytogenes egd-e.
|
| 61 | c3grzA_ |
|
not modelled |
33.4 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:ribosomal protein l11 methyltransferase;
PDBTitle: crystal structure of ribosomal protein l11 methylase from2 lactobacillus delbrueckii subsp. bulgaricus
|
| 62 | d2jgra1 |
|
not modelled |
32.8 |
19 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
| 63 | d1fp1d2 |
|
not modelled |
32.5 |
12 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 64 | d1iiba_ |
|
not modelled |
31.5 |
25 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 65 | c3gxaA_ |
|
not modelled |
30.8 |
30 |
PDB header:protein binding
Chain: A: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of gna1946
|
| 66 | c2jrlA_ |
|
not modelled |
30.7 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
|
| 67 | c2esrB_ |
|
not modelled |
30.7 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: conserved hypothetical protein- streptococcus pyogenes
|
| 68 | c3dzcA_ |
|
not modelled |
29.8 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: 2.35 angstrom resolution structure of wecb (vc0917), a udp-n-2 acetylglucosamine 2-epimerase from vibrio cholerae.
|
| 69 | c2p35A_ |
|
not modelled |
29.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:trans-aconitate 2-methyltransferase;
PDBTitle: crystal structure of trans-aconitate methyltransferase from2 agrobacterium tumefaciens
|
| 70 | c3kklA_ |
|
not modelled |
28.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable chaperone protein hsp33;
PDBTitle: crystal structure of functionally unknown hsp33 from2 saccharomyces cerevisiae
|
| 71 | c2i2xD_ |
|
not modelled |
28.6 |
35 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
| 72 | c3d4oA_ |
|
not modelled |
28.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 73 | d2naca2 |
|
not modelled |
28.2 |
13 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 74 | d1vhta_ |
|
not modelled |
28.0 |
26 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 75 | c3crnA_ |
|
not modelled |
27.9 |
24 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver domain protein, chey-like;
PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
|
| 76 | c1yr3A_ |
|
not modelled |
27.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:xanthosine phosphorylase;
PDBTitle: escherichia coli purine nucleoside phosphorylase ii, the2 product of the xapa gene
|
| 77 | d1fp2a2 |
|
not modelled |
27.0 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
| 78 | c3h5dD_ |
|
not modelled |
26.6 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
| 79 | c3clhA_ |
|
not modelled |
26.5 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of 3-dehydroquinate synthase (dhqs)from2 helicobacter pylori
|
| 80 | c2q1wC_ |
|
not modelled |
25.8 |
41 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
| 81 | c1y80A_ |
|
not modelled |
24.8 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 82 | d3pmga1 |
|
not modelled |
24.8 |
16 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 83 | d1mgta2 |
|
not modelled |
24.7 |
33 |
Fold:Ribonuclease H-like motif
Superfamily:Methylated DNA-protein cysteine methyltransferase domain
Family:Methylated DNA-protein cysteine methyltransferase domain |
| 84 | d1j8fa_ |
|
not modelled |
24.2 |
30 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 85 | d1qrda_ |
|
not modelled |
24.0 |
20 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 86 | c2rirA_ |
|
not modelled |
23.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 87 | c3nbmA_ |
|
not modelled |
22.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, lactose-specific iibc components;
PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
|
| 88 | c2i2aA_ |
|
not modelled |
22.1 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:probable inorganic polyphosphate/atp-nad kinase 1;
PDBTitle: crystal structure of lmnadk1 from listeria monocytogenes
|
| 89 | d1lrza3 |
|
not modelled |
21.9 |
10 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:FemXAB nonribosomal peptidyltransferases |
| 90 | c3g07C_ |
|
not modelled |
21.7 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:7sk snrna methylphosphate capping enzyme;
PDBTitle: methyltransferase domain of human bicoid-interacting protein2 3 homolog (drosophila)
|
| 91 | d1qe5a_ |
|
not modelled |
21.6 |
31 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 92 | c3i42A_ |
|
not modelled |
21.5 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:response regulator receiver domain protein (chey-
PDBTitle: structure of response regulator receiver domain (chey-like)2 from methylobacillus flagellatus
|
| 93 | c3nhzA_ |
|
not modelled |
21.4 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:two component system transcriptional regulator mtra;
PDBTitle: structure of n-terminal domain of mtra
|
| 94 | d2cara1 |
|
not modelled |
21.4 |
17 |
Fold:Anticodon-binding domain-like
Superfamily:ITPase-like
Family:ITPase (Ham1) |
| 95 | c3daqB_ |
|
not modelled |
21.3 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 96 | d2nxca1 |
|
not modelled |
20.7 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Ribosomal protein L11 methyltransferase PrmA |
| 97 | d1gpja2 |
|
not modelled |
20.7 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 98 | c3cfyA_ |
|
not modelled |
20.1 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative luxo repressor protein;
PDBTitle: crystal structure of signal receiver domain of putative luxo2 repressor protein from vibrio parahaemolyticus
|
| 99 | d1wd7a_ |
|
not modelled |
20.0 |
18 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |