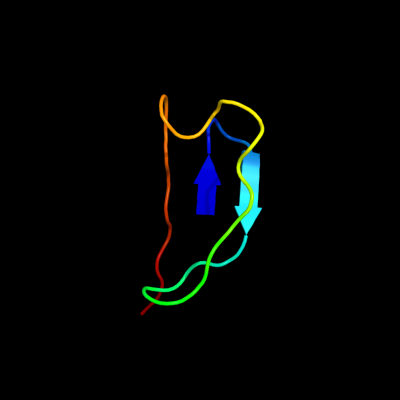
1 d1yq2a3
68.8
32
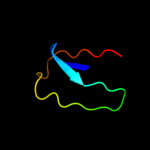
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain2 d1jz8a3
67.9
28
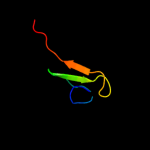
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain3 c1yj5C_
54.7
26
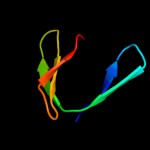
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
4 c3bgaB_
53.0
22
PDB header: hydrolaseChain: B: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of beta-galactosidase from bacteroides2 thetaiotaomicron vpi-5482
5 d2je8a4
52.4
15
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain6 c3qhsD_
50.0
23
PDB header: rna binding proteinChain: D: PDB Molecule: protein hfq;PDBTitle: crystal structure of full-length hfq from escherichia coli
7 d1hk9a_
49.9
23
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Pleiotropic translational regulator Hfq8 c3poaA_
47.6
21
PDB header: peptide binding proteinChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: structural and functional analysis of phosphothreonine-dependent fha2 domain interactions
9 d1u1sa1
47.4
23
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Pleiotropic translational regulator Hfq10 d1yjma1
43.5
26
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain11 c2je8B_
42.4
15
PDB header: hydrolaseChain: B: PDB Molecule: beta-mannosidase;PDBTitle: structure of a beta-mannosidase from bacteroides2 thetaiotaomicron
12 c1jz6C_
40.3
28
PDB header: hydrolaseChain: C: PDB Molecule: beta-galactosidase;PDBTitle: e. coli (lacz) beta-galactosidase in complex with galacto-2 tetrazole
13 c3obaA_
40.1
25
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: structure of the beta-galactosidase from kluyveromyces lactis
14 c1yq2C_
39.4
34
PDB header: hydrolaseChain: C: PDB Molecule: beta-galactosidase;PDBTitle: beta-galactosidase from arthrobacter sp. c2-2 (isoenzyme c2-2 2-1)
15 c3pe9B_
35.8
15
PDB header: unknown functionChain: B: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
16 c3mv14_
34.8
28
PDB header: hydrolaseChain: 4: PDB Molecule: beta-galactosidase;PDBTitle: e.coli (lacz) beta-galactosidase (r599a) in complex with guanidinium
17 d1ujxa_
32.4
25
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain18 d2vzsa4
25.9
26
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain19 d1tg7a2
25.5
3
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Beta-galactosidase LacA, domains 4 and 520 c3u7zA_
25.0
17
PDB header: metal binding proteinChain: A: PDB Molecule: putative metal binding protein rumgna_00854;PDBTitle: crystal structure of a putative metal binding protein rumgna_008542 (zp_02040092.1) from ruminococcus gnavus atcc 29149 at 1.30 a3 resolution
21 c3gm8A_
not modelled
22.4
15
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase family 2, candidate beta-glycosidase;PDBTitle: crystal structure of a beta-glycosidase from bacteroides vulgatus
22 c2qguA_
not modelled
21.6
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable signal peptide protein;PDBTitle: three-dimensional structure of the phospholipid-binding protein from2 ralstonia solanacearum q8xv73_ralsq in complex with a phospholipid at3 the resolution 1.53 a. northeast structural genomics consortium4 target rsr89
23 c2gtqA_
not modelled
21.3
15
PDB header: hydrolaseChain: A: PDB Molecule: aminopeptidase n;PDBTitle: crystal structure of aminopeptidase n from human pathogen neisseria2 meningitidis
24 d2nqda1
not modelled
19.2
7
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ICP-likeFamily: ICP-like25 c2h0eA_
not modelled
18.8
17
PDB header: hydrolaseChain: A: PDB Molecule: transthyretin-like protein pucm;PDBTitle: crystal structure of pucm in the absence of substrate
26 d1lgpa_
not modelled
18.1
24
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain27 d1bhga2
not modelled
17.9
13
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain28 d2jnaa1
not modelled
16.9
23
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like29 c2w1wB_
not modelled
16.1
13
PDB header: hydrolaseChain: B: PDB Molecule: lipolytic enzyme, g-d-s-l;PDBTitle: native structure of a family 35 carbohydrate binding module2 from clostridium thermocellum
30 d2c34a1
not modelled
16.0
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ICP-likeFamily: ICP-like31 d3bvua3
not modelled
15.2
0
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: alpha-mannosidase32 c3pe9C_
not modelled
15.0
10
PDB header: unknown functionChain: C: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
33 c2h1xB_
not modelled
13.8
18
PDB header: hydrolaseChain: B: PDB Molecule: 5-hydroxyisourate hydrolase (formerly known asPDBTitle: crystal structure of 5-hydroxyisourate hydrolase (formerly2 known as trp, transthyretin related protein)
34 c2gpzC_
not modelled
13.6
16
PDB header: hydrolaseChain: C: PDB Molecule: transthyretin-like protein;PDBTitle: transthyretin-like protein from salmonella dublin
35 c3pe9A_
not modelled
13.6
10
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
36 c1htyA_
not modelled
13.6
0
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase ii;PDBTitle: golgi alpha-mannosidase ii
37 d1w9sa_
not modelled
13.2
3
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM638 d1oo2a_
not modelled
13.2
9
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)39 d2g1la1
not modelled
12.7
33
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain40 c3ff6D_
not modelled
12.6
32
PDB header: ligaseChain: D: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: human acc2 ct domain with cp-640186
41 c3qvaB_
not modelled
12.5
11
PDB header: hydrolaseChain: B: PDB Molecule: transthyretin-like protein;PDBTitle: structure of klebsiella pneumoniae 5-hydroxyisourate hydrolase
42 d1ttaa_
not modelled
12.2
9
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)43 c3htxA_
not modelled
11.9
64
PDB header: transferase/rnaChain: A: PDB Molecule: hen1;PDBTitle: crystal structure of small rna methyltransferase hen1
44 c2x24B_
not modelled
11.8
28
PDB header: ligaseChain: B: PDB Molecule: acetyl-coa carboxylase;PDBTitle: bovine acc2 ct domain in complex with inhibitor
45 c2ow7A_
not modelled
11.8
0
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase 2;PDBTitle: golgi alpha-mannosidase ii complex with (1r,6s,7r,8s)-1-2 thioniabicyclo[4.3.0]nonan-7,8-diol chloride
46 d2oeza1
not modelled
11.7
27
Fold: YacF-likeSuperfamily: YacF-likeFamily: YacF-like47 c3u2bC_
not modelled
11.5
27
PDB header: transcription/dnaChain: C: PDB Molecule: transcription factor sox-4;PDBTitle: structure of the sox4 hmg domain bound to dna
48 c2wgnB_
not modelled
11.5
7
PDB header: hydrolase inhibitorChain: B: PDB Molecule: inhibitor of cysteine peptidase compnd 3;PDBTitle: pseudomonas aeruginosa icp
49 c3hx1B_
not modelled
11.2
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1951 protein;PDBTitle: crystal structure of the slr1951 protein from synechocystis sp.2 northeast structural genomics consortium target sgr167a
50 c2v4vA_
not modelled
11.0
12
PDB header: hydrolaseChain: A: PDB Molecule: gh59 galactosidase;PDBTitle: crystal structure of a family 6 carbohydrate-binding module2 from clostridium cellulolyticum in complex with xylose
51 d1od3a_
not modelled
10.7
14
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM652 d1i11a_
not modelled
10.6
9
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box53 c3fm8A_
not modelled
10.5
30
PDB header: transport protein/hydrolase activatorChain: A: PDB Molecule: kinesin-like protein kif13b;PDBTitle: crystal structure of full length centaurin alpha-1 bound with the fha2 domain of kif13b (capri target)
54 d1kq1a_
not modelled
10.4
14
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Pleiotropic translational regulator Hfq55 d1uy4a_
not modelled
10.2
18
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM656 c3cmgA_
not modelled
9.7
12
PDB header: hydrolaseChain: A: PDB Molecule: putative beta-galactosidase;PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
57 d2g82a1
not modelled
9.6
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain58 d1f86a_
not modelled
9.5
9
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)59 c3d0wD_
not modelled
9.4
24
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: yflh protein;PDBTitle: crystal structure of yflh protein from bacillus subtilis.2 northeast structural genomics consortium target sr326
60 c3lpgA_
not modelled
8.9
8
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: beta-glucuronidase;PDBTitle: structure of e. coli beta-glucuronidase bound with a novel, potent2 inhibitor 3-(2-fluorophenyl)-1-(2-hydroxyethyl)-1-((6-methyl-2-oxo-1,3 2-dihydroquinolin-3-yl)methyl)urea
61 c3kcmC_
not modelled
8.6
13
PDB header: oxidoreductaseChain: C: PDB Molecule: thioredoxin family protein;PDBTitle: the crystal structure of thioredoxin protein from geobacter2 metallireducens
62 d1tfpa_
not modelled
8.6
9
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)63 d2d6fc3
not modelled
8.6
9
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: GatB/GatE catalytic domain-like64 c2crjA_
not modelled
8.6
21
PDB header: gene regulationChain: A: PDB Molecule: swi/snf-related matrix-associated actin-PDBTitle: solution structure of the hmg domain of mouse hmg domain2 protein hmgx2
65 c3pe9D_
not modelled
8.2
10
PDB header: unknown functionChain: D: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
66 d1sr3a_
not modelled
8.1
14
Fold: OB-foldSuperfamily: Heme chaperone CcmEFamily: Heme chaperone CcmE67 d1hsma_
not modelled
7.9
15
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box68 d1wlna1
not modelled
7.8
20
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain69 c1d0rA_
not modelled
7.8
33
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
70 d1uxxx_
not modelled
7.8
23
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM671 c2co9A_
not modelled
7.7
6
PDB header: transcriptionChain: A: PDB Molecule: thymus high mobility group box protein tox;PDBTitle: solution structure of the hmg_box domain of thymus high2 mobility group box protein tox from mouse
72 d1n67a1
not modelled
7.4
10
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Fibrinogen-binding domain73 c2bgpA_
not modelled
7.4
27
PDB header: carbohydrate binding proteinChain: A: PDB Molecule: endo-b1,4-mannanase 5c;PDBTitle: mannan binding module from man5c in bound conformation
74 d1k1fa_
not modelled
7.3
13
Fold: Bcr-Abl oncoprotein oligomerization domainSuperfamily: Bcr-Abl oncoprotein oligomerization domainFamily: Bcr-Abl oncoprotein oligomerization domain75 c1yq1A_
not modelled
7.3
14
PDB header: transferaseChain: A: PDB Molecule: glutathione s-transferase;PDBTitle: structural genomics of caenorhabditis elegans: glutathione2 s-transferase
76 c1hrzA_
not modelled
7.2
24
PDB header: dna binding protein/dnaChain: A: PDB Molecule: human sry;PDBTitle: the 3d structure of the human sry-dna complex solved by2 multi-dimensional heteronuclear-edited and-filtered nmr
77 d2gzka2
not modelled
7.2
24
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box78 c1hryA_
not modelled
7.2
24
PDB header: dna binding protein/dnaChain: A: PDB Molecule: human sry;PDBTitle: the 3d structure of the human sry-dna complex solved by2 multid-dimensional heteronuclear-edited and-filtered nmr
79 c2vzvB_
not modelled
7.2
28
PDB header: hydrolaseChain: B: PDB Molecule: exo-beta-d-glucosaminidase;PDBTitle: substrate complex of amycolatopsis orientalis exo-2 chitosanase csxa e541a with chitosan
80 d2noca1
not modelled
7.1
39
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like81 c2cdpC_
not modelled
7.0
29
PDB header: hydrolaseChain: C: PDB Molecule: beta-agarase 1;PDBTitle: structure of a cbm6 in complex with neoagarohexaose
82 d1uxza_
not modelled
6.9
24
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM683 c1hmfA_
not modelled
6.8
15
PDB header: dna-bindingChain: A: PDB Molecule: high mobility group protein fragment-b;PDBTitle: structure of the hmg box motif in the b-domain of hmg1
84 d1hmfa_
not modelled
6.8
15
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box85 d1kgia_
not modelled
6.8
7
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)86 c3kzeA_
not modelled
6.8
12
PDB header: signaling proteinChain: A: PDB Molecule: t-lymphoma invasion and metastasis-inducing protein 1;PDBTitle: crystal structure of t-cell lymphoma invasion and metastasis-1 pdz in2 complex with ssrkeyya peptide
87 c1z5hB_
not modelled
6.7
12
PDB header: hydrolaseChain: B: PDB Molecule: tricorn protease interacting factor f3;PDBTitle: crystal structures of the tricorn interacting factor f32 from thermoplasma acidophilum
88 c2kklA_
not modelled
6.7
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mb1858;PDBTitle: solution nmr structure of fha domain of mb1858 from2 mycobacterium bovis. northeast structural genomics3 consortium target mbr243c (24-155).
89 d1z0mb1
not modelled
6.7
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like90 d1zzoa1
not modelled
6.6
13
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like91 c3hsbB_
not modelled
6.6
17
PDB header: rna binding protein/rnaChain: B: PDB Molecule: protein hfq;PDBTitle: crystal structure of ymah (hfq) from bacillus subtilis in complex with2 an rna aptamer
92 c3k1rA_
not modelled
6.6
19
PDB header: structural proteinChain: A: PDB Molecule: harmonin;PDBTitle: structure of harmonin npdz1 in complex with the sam-pbm of2 sans
93 c1kq1W_
not modelled
6.4
14
PDB header: translationChain: W: PDB Molecule: host factor for q beta;PDBTitle: 1.55 a crystal structure of the pleiotropic translational2 regulator, hfq
94 c3cynC_
not modelled
6.3
14
PDB header: oxidoreductaseChain: C: PDB Molecule: probable glutathione peroxidase 8;PDBTitle: the structure of human gpx8
95 c2vr3B_
not modelled
6.3
10
PDB header: cell adhesionChain: B: PDB Molecule: clumping factor a;PDBTitle: structural and biochemical characterization of fibrinogen2 binding to clfa from staphylocccus aureus
96 c2vzqA_
not modelled
6.3
13
PDB header: hydrolaseChain: A: PDB Molecule: exo-beta-d-glucosaminidase;PDBTitle: c-terminal cbm35 from amycolatopsis orientalis exo-2 chitosanase csxa in complex with digalacturonic acid
97 c2p5qA_
not modelled
6.2
7
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase 5;PDBTitle: crystal structure of the poplar glutathione peroxidase 5 in2 the reduced form
98 d2ff4a3
not modelled
6.2
20
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain99 d1j3da_
not modelled
6.2
21
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box