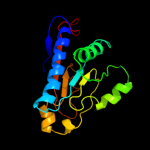
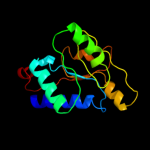
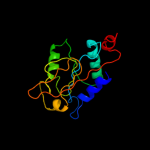
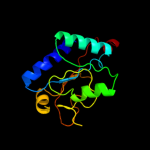
1 c1hf2A_
100.0
23
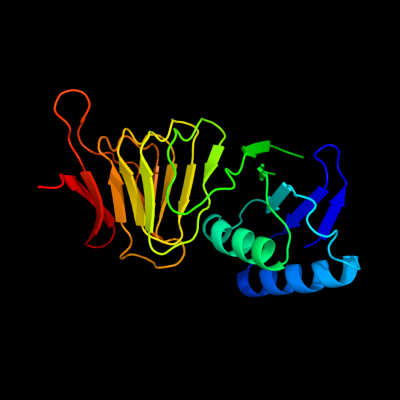
PDB header: cell division proteinChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the bacterial cell-division inhibitor2 minc from t. maritima
2 d1hf2a1
100.0
27
Fold: Single-stranded right-handed beta-helixSuperfamily: Cell-division inhibitor MinC, C-terminal domainFamily: Cell-division inhibitor MinC, C-terminal domain3 c3ghfA_
99.8
83
PDB header: cell cycleChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the septum site-determining protein2 minc from salmonella typhimurium
4 d1hf2a2
98.5
16
Fold: Cell-division inhibitor MinC, N-terminal domainSuperfamily: Cell-division inhibitor MinC, N-terminal domainFamily: Cell-division inhibitor MinC, N-terminal domain5 c1tf2A_
90.6
25
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of seca:adp in an open conformation from2 bacillus subtilis
6 c1nl3B_
86.7
27
PDB header: protein transportChain: B: PDB Molecule: preprotein translocase seca 1 subunit;PDBTitle: crystal structure of the seca protein translocation atpase2 from mycobacterium tuberculosis in apo form
7 c3dl8B_
84.1
22
PDB header: protein transportChain: B: PDB Molecule: protein translocase subunit seca;PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
8 c3dinB_
83.0
25
PDB header: membrane protein, protein transportChain: B: PDB Molecule: protein translocase subunit seca;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
9 c3juxA_
82.2
25
PDB header: protein transportChain: A: PDB Molecule: protein translocase subunit seca;PDBTitle: structure of the translocation atpase seca from thermotoga2 maritima
10 c2fsgA_
81.9
25
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase seca subunit;PDBTitle: complex seca:atp from escherichia coli
11 d1nkta4
79.0
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain12 d1mzva_
75.7
15
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)13 c2vdaA_
74.2
26
PDB header: protein transportChain: A: PDB Molecule: translocase subunit seca;PDBTitle: solution structure of the seca-signal peptide complex
14 c2dy0A_
65.4
13
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of project jw0458 from escherichia coli
15 c2pfuA_
61.4
12
PDB header: transport proteinChain: A: PDB Molecule: biopolymer transport exbd protein;PDBTitle: nmr strcuture determination of the periplasmic domain of exbd from2 e.coli
16 c2ipcB_
53.1
29
PDB header: transport proteinChain: B: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of the translocation atpase seca from thermus2 thermophilus reveals a parallel, head-to-head dimer
17 d1zunb1
41.3
18
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors18 c3l7nA_
40.1
8
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of smu.1228c
19 d1g2qa_
37.6
13
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)20 d1o57a2
37.1
13
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)21 d1n0ua1
not modelled
36.8
33
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors22 c2jwlB_
not modelled
33.3
6
PDB header: membrane proteinChain: B: PDB Molecule: protein tolr;PDBTitle: solution structure of periplasmic domain of tolr from h.2 influenzae with saxs data
23 c3k4iC_
not modelled
31.0
20
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein pspto_3204 from2 pseudomonas syringae pv. tomato str. dc3000
24 d2bv3a1
not modelled
27.3
21
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors25 d1jnya1
not modelled
26.4
15
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors26 c2i5kB_
not modelled
26.3
19
PDB header: transferaseChain: B: PDB Molecule: utp--glucose-1-phosphate uridylyltransferase;PDBTitle: crystal structure of ugp1p
27 d1o1ya_
not modelled
24.1
10
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)28 c2zwaA_
not modelled
23.9
17
PDB header: transferaseChain: A: PDB Molecule: leucine carboxyl methyltransferase 2;PDBTitle: crystal structure of trna wybutosine synthesizing enzyme2 tyw4
29 c3cr8C_
not modelled
23.9
8
PDB header: transferaseChain: C: PDB Molecule: sulfate adenylyltranferase, adenylylsulfatePDBTitle: hexameric aps kinase from thiobacillus denitrificans
30 d1l1qa_
not modelled
22.4
12
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)31 d1qb7a_
not modelled
21.9
15
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)32 d2f69a1
not modelled
21.4
29
Fold: open-sided beta-meanderSuperfamily: Histone H3 K4-specific methyltransferase SET7/9 N-terminal domainFamily: Histone H3 K4-specific methyltransferase SET7/9 N-terminal domain33 d1f60a1
not modelled
20.0
12
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors34 c2idcA_
not modelled
19.9
17
PDB header: replication/chaperoneChain: A: PDB Molecule: anti-silencing protein 1 and histone h3 chimera;PDBTitle: structure of the histone h3-asf1 chaperone interaction
35 d1roca_
not modelled
18.8
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ASF1-likeFamily: ASF1-like36 d2icya1
not modelled
17.9
24
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: GlmU C-terminal domain-like37 c2i32A_
not modelled
17.8
22
PDB header: replication chaperoneChain: A: PDB Molecule: anti-silencing factor 1 paralog a;PDBTitle: structure of a human asf1a-hira complex and insights into2 specificity of histone chaperone complex assembly
38 d2i32a1
not modelled
17.8
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ASF1-likeFamily: ASF1-like39 d2c78a1
not modelled
16.7
10
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors40 d1v30a_
not modelled
16.6
56
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like41 c3es1A_
not modelled
16.6
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of protein with a cupin-like fold and unknown2 function (yp_001165807.1) from novosphingobium aromaticivorans dsm3 12444 at 1.91 a resolution
42 d1mvfd_
not modelled
16.6
22
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: Kis/PemI addiction antidote43 d2exda1
not modelled
16.6
18
Fold: OB-foldSuperfamily: NfeD domain-likeFamily: NfeD domain-like44 c3agqA_
not modelled
16.5
9
PDB header: translation,transferaseChain: A: PDB Molecule: elongation factor ts, elongation factor tu 1, linker, qPDBTitle: structure of viral polymerase form ii
45 c3tr5C_
not modelled
16.4
18
PDB header: translationChain: C: PDB Molecule: peptide chain release factor 3;PDBTitle: structure of a peptide chain release factor 3 (prfc) from coxiella2 burnetii
46 d2cu9a1
not modelled
16.1
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ASF1-likeFamily: ASF1-like47 c2eqaA_
not modelled
14.7
18
PDB header: rna binding proteinChain: A: PDB Molecule: hypothetical protein st1526;PDBTitle: crystal structure of the hypothetical sua5 protein from2 sulfolobus tokodaii
48 c3k13A_
not modelled
14.1
17
PDB header: transferaseChain: A: PDB Molecule: 5-methyltetrahydrofolate-homocysteine methyltransferase;PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
49 d1vkba_
not modelled
14.0
44
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like50 c2h5eB_
not modelled
13.7
16
PDB header: translationChain: B: PDB Molecule: peptide chain release factor rf-3;PDBTitle: crystal structure of e.coli polypeptide release factor rf3
51 d1q1oa_
not modelled
13.5
10
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain52 c2wmkB_
not modelled
13.3
14
PDB header: hydrolaseChain: B: PDB Molecule: fucolectin-related protein;PDBTitle: crystal structure of the catalytic module of a family 982 glycoside hydrolase from streptococcus pneumoniae sp3-bs713 (sp3gh98) in complex with the a-lewisy pentasaccharide4 blood group antigen.
53 c2xexA_
not modelled
13.1
13
PDB header: translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of staphylococcus aureus elongation factor2 g
54 d1dcsa_
not modelled
13.0
16
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Penicillin synthase-like55 c3mezA_
not modelled
12.7
15
PDB header: sugar binding proteinChain: A: PDB Molecule: mannose-specific lectin 3 chain 1;PDBTitle: x-ray structural analysis of a mannose specific lectin from dutch2 crocus (crocus vernus)
56 c2kl8A_
not modelled
12.2
23
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
57 d1iqoa_
not modelled
12.2
21
Fold: Hypothetical protein MTH1880Superfamily: Hypothetical protein MTH1880Family: Hypothetical protein MTH188058 d1pqsa_
not modelled
11.8
8
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain59 c3l83A_
not modelled
11.7
15
PDB header: transferaseChain: A: PDB Molecule: glutamine amido transferase;PDBTitle: crystal structure of glutamine amido transferase from methylobacillus2 flagellatus
60 c3jubA_
not modelled
11.3
38
PDB header: transferaseChain: A: PDB Molecule: aig2-like domain-containing protein 1;PDBTitle: human gamma-glutamylamine cyclotransferase
61 d1omwa2
not modelled
11.1
15
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)62 c2rdo7_
not modelled
10.8
13
PDB header: ribosomeChain: 7: PDB Molecule: elongation factor g;PDBTitle: 50s subunit with ef-g(gdpnp) and rrf bound
63 c2k5hA_
not modelled
10.5
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein;PDBTitle: solution nmr structure of protein encoded by mth693 from2 methanobacterium thermoautotrophicum: northeast structural3 genomics consortium target tt824a
64 d1vm6a3
not modelled
10.2
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain65 c3uotB_
not modelled
10.0
13
PDB header: cell cycleChain: B: PDB Molecule: mediator of dna damage checkpoint protein 1;PDBTitle: crystal structure of mdc1 fha domain in complex with a phosphorylated2 peptide from the mdc1 n-terminus
66 c3cb4D_
not modelled
10.0
14
PDB header: translationChain: D: PDB Molecule: gtp-binding protein lepa;PDBTitle: the crystal structure of lepa
67 d1xhsa_
not modelled
10.0
56
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like68 c2g0qA_
not modelled
9.8
44
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: at5g39720.1 protein;PDBTitle: solution structure of at5g39720.1 from arabidopsis thaliana
69 c2vu9A_
not modelled
9.6
16
PDB header: hydrolaseChain: A: PDB Molecule: botulinum neurotoxin a heavy chain;PDBTitle: crystal structure of botulinum neurotoxin serotype a2 binding domain in complex with gt1b
70 c2bm0A_
not modelled
9.5
15
PDB header: elongation factorChain: A: PDB Molecule: elongation factor g;PDBTitle: ribosomal elongation factor g (ef-g) fusidic acid resistant2 mutant t84a
71 c1qysA_
not modelled
9.4
21
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
72 d1od5a2
not modelled
9.2
21
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein73 d2cvoa2
not modelled
9.1
16
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like74 c3k3sG_
not modelled
8.9
17
PDB header: hydrolaseChain: G: PDB Molecule: altronate hydrolase;PDBTitle: crystal structure of altronate hydrolase (fragment 1-84) from shigella2 flexneri.
75 c2q4jB_
not modelled
8.9
16
PDB header: transferaseChain: B: PDB Molecule: probable utp-glucose-1-phosphate uridylyltransferase 2;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at3g03250, a putative udp-glucose3 pyrophosphorylase
76 c2jqvA_
not modelled
8.8
44
PDB header: structural genomicsChain: A: PDB Molecule: aig2 protein-like;PDBTitle: solution structure at3g28950.1 from arabidopsis thaliana
77 c3cgiD_
not modelled
8.8
36
PDB header: unknown functionChain: D: PDB Molecule: propanediol utilization protein pduu;PDBTitle: crystal structure of the pduu shell protein from the pdu2 microcompartment
78 c3mmyE_
not modelled
8.8
16
PDB header: nuclear proteinChain: E: PDB Molecule: mrna export factor;PDBTitle: structural and functional analysis of the interaction between the2 nucleoporin nup98 and the mrna export factor rae1
79 d1ip9a_
not modelled
8.7
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain80 c2d5nB_
not modelled
8.7
18
PDB header: hydrolase, oxidoreductaseChain: B: PDB Molecule: riboflavin biosynthesis protein ribd;PDBTitle: crystal structure of a bifunctional deaminase and reductase2 involved in riboflavin biosynthesis
81 c2qikA_
not modelled
8.7
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0131 protein ykqa;PDBTitle: crystal structure of ykqa from bacillus subtilis. northeast2 structural genomics target sr631
82 d1ofda1
not modelled
8.7
14
Fold: Single-stranded right-handed beta-helixSuperfamily: Alpha subunit of glutamate synthase, C-terminal domainFamily: Alpha subunit of glutamate synthase, C-terminal domain83 c2jvfA_
not modelled
8.7
18
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
84 c2glvA_
not modelled
8.6
15
PDB header: lyaseChain: A: PDB Molecule: (3r)-hydroxymyristoyl-acyl carrier proteinPDBTitle: crystal structure of (3r)-hydroxyacyl-acyl carrier protein2 dehydratase(fabz) mutant(y100a) from helicobacter pylori
85 c3r0eC_
not modelled
8.5
36
PDB header: sugar binding proteinChain: C: PDB Molecule: lectin;PDBTitle: structure of remusatia vivipara lectin
86 c2qv3A_
not modelled
8.2
57
PDB header: toxinChain: A: PDB Molecule: vacuolating cytotoxin;PDBTitle: crystal structure of the helicobacter pylori vacuolating toxin p552 domain
87 d2dy1a1
not modelled
7.7
16
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors88 c3b8hA_
not modelled
7.7
24
PDB header: biosynthetic protein/transferaseChain: A: PDB Molecule: elongation factor 2;PDBTitle: structure of the eef2-exoa(e546a)-nad+ complex
89 c3ia0c_
not modelled
7.6
27
PDB header: structural proteinChain: C: PDB Molecule: ethanolamine utilization protein euts;PDBTitle: ethanolamine utilization microcompartment shell subunit,2 euts-g39v mutant
90 c2jw5A_
not modelled
7.5
12
PDB header: protein bindingChain: A: PDB Molecule: dna polymerase lambda;PDBTitle: polymerase lambda brct domain
91 d1y13a_
not modelled
7.5
19
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: 6-pyruvoyl tetrahydropterin synthase92 c3jx9B_
not modelled
7.4
18
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of putative phosphoheptose isomerase2 (yp_001815198.1) from exiguobacterium sp. 255-15 at 1.95 a resolution
93 c1g2cN_
not modelled
7.4
22
PDB header: viral proteinChain: N: PDB Molecule: fusion protein (f);PDBTitle: human respiratory syncytial virus fusion protein core
94 d2gsaa_
not modelled
7.4
3
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like95 c2w18A_
not modelled
7.1
30
PDB header: nuclear proteinChain: A: PDB Molecule: partner and localizer of brca2;PDBTitle: crystal structure of the c-terminal wd40 domain of human2 palb2
96 d1diha1
not modelled
7.1
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain97 d1d9ea_
not modelled
7.0
9
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase98 c3mkqA_
not modelled
7.0
9
PDB header: transport proteinChain: A: PDB Molecule: coatomer beta'-subunit;PDBTitle: crystal structure of yeast alpha/betaprime-cop subcomplex of the copi2 vesicular coat
99 d1fxza2
not modelled
7.0
18
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein