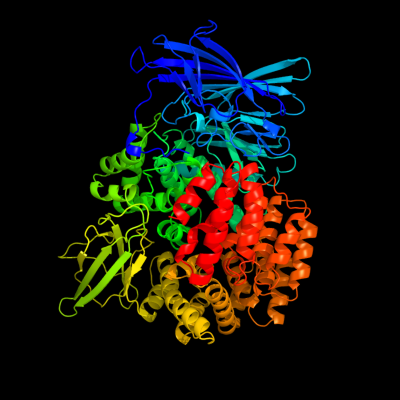
| 1 | c3b37A_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:aminopeptidase n;
PDBTitle: crystal structure of e. coli aminopeptidase n in complex with tyrosine
|
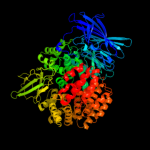
| 2 | c3ebhA_
|
|
 |
100.0 |
35 |
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:m1 family aminopeptidase;
PDBTitle: structure of the m1 alanylaminopeptidase from malaria complexed with2 bestatin
|
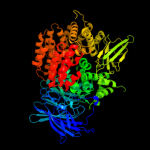
| 3 | c2gtqA_
|
|
 |
100.0 |
47 |
PDB header:hydrolase
Chain: A: PDB Molecule:aminopeptidase n;
PDBTitle: crystal structure of aminopeptidase n from human pathogen neisseria2 meningitidis
|
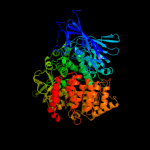
| 4 | c3se6A_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoplasmic reticulum aminopeptidase 2;
PDBTitle: crystal structure of the human endoplasmic reticulum aminopeptidase 2
|
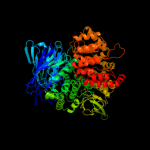
| 5 | c2xdtA_
|
|
 |
100.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoplasmic reticulum aminopeptidase 1;
PDBTitle: crystal structure of the soluble domain of human2 endoplasmic reticulum aminopeptidase 1 erap1
|
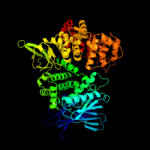
| 6 | c3mdjB_
|
|
 |
100.0 |
17 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:endoplasmic reticulum aminopeptidase 1;
PDBTitle: er aminopeptidase, erap1, bound to the zinc aminopeptidase inhibitor,2 bestatin
|
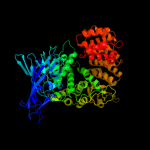
| 7 | c1z5hB_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:tricorn protease interacting factor f3;
PDBTitle: crystal structures of the tricorn interacting factor f32 from thermoplasma acidophilum
|
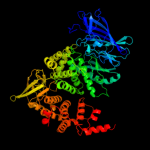
| 8 | c3qnfA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoplasmic reticulum aminopeptidase 1;
PDBTitle: crystal structure of the open state of human endoplasmic reticulum2 aminopeptidase 1 erap1
|
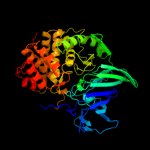
| 9 | c3b7uX_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: X: PDB Molecule:leukotriene a-4 hydrolase;
PDBTitle: leukotriene a4 hydrolase complexed with kelatorphan
|
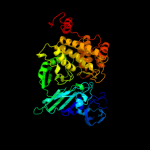
| 10 | c3ciaA_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:cold-active aminopeptidase;
PDBTitle: crystal structure of cold-aminopeptidase from colwellia2 psychrerythraea
|
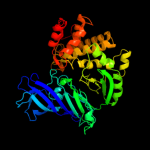
| 11 | c2xpyA_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:leukotriene a-4 hydrolase;
PDBTitle: structure of native leukotriene a4 hydrolase from saccharomyces2 cerevisiae
|
| 12 | d3b7sa3
|
|
 |
100.0 |
24 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Leukotriene A4 hydrolase catalytic domain |
| 13 | d3b7sa2
|
|
 |
100.0 |
23 |
Fold:Leukotriene A4 hydrolase N-terminal domain
Superfamily:Leukotriene A4 hydrolase N-terminal domain
Family:Leukotriene A4 hydrolase N-terminal domain |
| 14 | c3rjoA_
|
|
 |
99.9 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoplasmic reticulum aminopeptidase 1;
PDBTitle: crystal structure of erap1 peptide binding domain
|
| 15 | c2vqxA_
|
|
 |
95.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:metalloproteinase;
PDBTitle: precursor of protealysin, metalloproteinase from serratia2 proteamaculans.
|
| 16 | d1u4ga_
|
|
 |
95.1 |
11 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 17 | d1npca_
|
|
 |
93.7 |
17 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 18 | c3cqbB_
|
|
 |
93.3 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable protease htpx homolog;
PDBTitle: crystal structure of heat shock protein htpx domain from vibrio2 parahaemolyticus rimd 2210633
|
| 19 | c3nqzB_
|
|
 |
91.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:secreted metalloprotease mcp02;
PDBTitle: crystal structure of the autoprocessed vibriolysin mcp-02 with e369a2 mutation
|
| 20 | c3nqxA_
|
|
 |
91.1 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:secreted metalloprotease mcp02;
PDBTitle: crystal structure of vibriolysin mcp-02 mature enzyme, a zinc2 metalloprotease from m4 family
|
| 21 | c3k7nA_ |
|
not modelled |
89.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:k-like;
PDBTitle: structures of two elapid snake venom metalloproteases with2 distinct activities highlight the disulfide patterns in the3 d domain of adamalysin family proteins
|
| 22 | d1bqba_ |
|
not modelled |
88.6 |
17 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 23 | c3k7lA_ |
|
not modelled |
88.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:atragin;
PDBTitle: structures of two elapid snake venom metalloproteases with2 distinct activities highlight the disulfide patterns in the3 d domain of adamalysin family proteins
|
| 24 | c2dw1B_ |
|
not modelled |
87.5 |
19 |
PDB header:apoptosis, toxin
Chain: B: PDB Molecule:catrocollastatin;
PDBTitle: crystal structure of vap2 from crotalus atrox venom (form 2-2 crystal)
|
| 25 | c2erpA_ |
|
not modelled |
86.1 |
17 |
PDB header:toxin
Chain: A: PDB Molecule:vascular apoptosis-inducing protein 1;
PDBTitle: crystal structure of vascular apoptosis-inducing protein-1(inhibitor-2 bound form)
|
| 26 | d1kjpa_ |
|
not modelled |
86.0 |
14 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 27 | d1r55a_ |
|
not modelled |
84.4 |
14 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 28 | c3c37B_ |
|
not modelled |
83.9 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidase, m48 family;
PDBTitle: x-ray structure of the putative zn-dependent peptidase q74d82 at the2 resolution 1.7 a. northeast structural genomics consortium target3 gsr143a
|
| 29 | d4aiga_ |
|
not modelled |
83.8 |
14 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 30 | c3g5cA_ |
|
not modelled |
82.1 |
14 |
PDB header:membrane protein
Chain: A: PDB Molecule:adam 22;
PDBTitle: structural and biochemical studies on the ectodomain of human adam22
|
| 31 | c2e3xA_ |
|
not modelled |
82.1 |
15 |
PDB header:hydrolase, blood clotting, toxin
Chain: A: PDB Molecule:coagulation factor x-activating enzyme heavy chain;
PDBTitle: crystal structure of russell's viper venom metalloproteinase
|
| 32 | d1nd1a_ |
|
not modelled |
79.4 |
16 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 33 | c2rjpC_ |
|
not modelled |
78.5 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:adamts-4;
PDBTitle: crystal structure of adamts4 with inhibitor bound
|
| 34 | d1quaa_ |
|
not modelled |
78.1 |
16 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 35 | d1kufa_ |
|
not modelled |
77.7 |
13 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 36 | d1atla_ |
|
not modelled |
77.5 |
13 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 37 | d1bswa_ |
|
not modelled |
77.3 |
16 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 38 | c2rjqA_ |
|
not modelled |
76.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:adamts-5;
PDBTitle: crystal structure of adamts5 with inhibitor bound
|
| 39 | c2y50A_ |
|
not modelled |
75.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:collagenase;
PDBTitle: crystal structure of collagenase g from clostridium2 histolyticum at 2.80 angstrom resolution
|
| 40 | d1wnia_ |
|
not modelled |
73.4 |
11 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 41 | c2j83B_ |
|
not modelled |
71.8 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:ulilysin;
PDBTitle: ulilysin metalloprotease in complex with batimastat.
|
| 42 | c1yp1A_ |
|
not modelled |
70.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:fii;
PDBTitle: crystal structure of a non-hemorrhagic fibrin(ogen)olytic2 metalloproteinase from venom of agkistrodon acutus
|
| 43 | c3iukB_ |
|
not modelled |
70.2 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of putative bacterial protein of unknown function2 (duf885, pf05960.1, ) from arthrobacter aurescens tc1, reveals fold3 similar to that of m32 carboxypeptidases
|
| 44 | c2v4bB_ |
|
not modelled |
69.1 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:adamts-1;
PDBTitle: crystal structure of human adamts-1 catalytic domain and2 cysteine-rich domain (apo-form)
|
| 45 | c3dtkA_ |
|
not modelled |
68.7 |
15 |
PDB header:gene regulation
Chain: A: PDB Molecule:irre protein;
PDBTitle: crystal structure of the irre protein, a central regulator2 of dna damage repair in deinococcaceae
|
| 46 | c3o0yC_ |
|
not modelled |
67.4 |
10 |
PDB header:lipid binding protein
Chain: C: PDB Molecule:lipoprotein;
PDBTitle: the crystal structure of the putative lipoprotein from colwellia2 psychrerythraea
|
| 47 | c2i47A_ |
|
not modelled |
61.9 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:adam 17;
PDBTitle: crystal structure of catalytic domain of tace with inhibitor
|
| 48 | c3lmcA_ |
|
not modelled |
61.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidase, zinc-dependent;
PDBTitle: crystal structure of zinc-dependent peptidase from methanocorpusculum2 labreanum (strain z), northeast structural genomics consortium target3 mur16
|
| 49 | c3b8zB_ |
|
not modelled |
61.1 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein adamts-5;
PDBTitle: high resolution crystal structure of the catalytic domain2 of adamts-5 (aggrecanase-2)
|
| 50 | d2i47a1 |
|
not modelled |
60.7 |
38 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:TNF-alpha converting enzyme, TACE, catalytic domain |
| 51 | c2qn0A_ |
|
not modelled |
59.2 |
21 |
PDB header:toxin
Chain: A: PDB Molecule:neurotoxin;
PDBTitle: structure of botulinum neurotoxin serotype c1 light chain2 protease
|
| 52 | c2xhqA_ |
|
not modelled |
57.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:archaemetzincin;
PDBTitle: crystal structure of archaemetzincin (amza) from archaeoglobus2 fulgidus at 1.45 a resolution
|
| 53 | c1z7hA_ |
|
not modelled |
57.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:tetanus toxin light chain;
PDBTitle: 2.3 angstrom crystal structure of tetanus neurotoxin light2 chain
|
| 54 | d1f83a_ |
|
not modelled |
56.8 |
26 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Clostridium neurotoxins, catalytic domain |
| 55 | d3btaa3 |
|
not modelled |
56.4 |
18 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Clostridium neurotoxins, catalytic domain |
| 56 | c3hjlA_ |
|
not modelled |
54.0 |
15 |
PDB header:proton transport
Chain: A: PDB Molecule:flagellar motor switch protein flig;
PDBTitle: the structure of full-length flig from aquifex aeolicus
|
| 57 | d1k9xa_ |
|
not modelled |
53.9 |
10 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Neurolysin-like |
| 58 | c3ka1A_ |
|
not modelled |
53.4 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:rbcx protein;
PDBTitle: crystal structure of rbcx from thermosynechococcus elongatus
|
| 59 | d1epwa3 |
|
not modelled |
52.9 |
26 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Clostridium neurotoxins, catalytic domain |
| 60 | d2ejqa1 |
|
not modelled |
49.4 |
6 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:TTHA0227-like |
| 61 | c2o0cB_ |
|
not modelled |
49.4 |
18 |
PDB header:signaling protein
Chain: B: PDB Molecule:alr2278 protein;
PDBTitle: crystal structure of the h-nox domain from nostoc sp. pcc 71202 complexed to no
|
| 62 | c3zusD_ |
|
not modelled |
49.0 |
18 |
PDB header:hydrolase/signaling protein
Chain: D: PDB Molecule:botulinum neurotoxin type a, synaptosomal-associated
PDBTitle: crystal structure of an engineered botulinum neurotoxin2 type a-snare23 derivative, lc-a-snap23-hn-a
|
| 63 | c2nyyA_ |
|
not modelled |
48.9 |
18 |
PDB header:toxin/immune system
Chain: A: PDB Molecule:botulinum neurotoxin type a;
PDBTitle: crystal structure of botulinum neurotoxin type a complexed with2 monoclonal antibody cr1
|
| 64 | d3bona1 |
|
not modelled |
47.4 |
18 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Clostridium neurotoxins, catalytic domain |
| 65 | c3deeA_ |
|
not modelled |
47.4 |
0 |
PDB header:transcription
Chain: A: PDB Molecule:putative regulatory protein;
PDBTitle: crystal structure of a putative regulatory protein involved in2 transcription (ngo1945) from neisseria gonorrhoeae fa 1090 at 2.25 a3 resolution
|
| 66 | d1j7na2 |
|
not modelled |
47.4 |
17 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Anthrax toxin lethal factor, N- and C-terminal domains |
| 67 | c3zuqA_ |
|
not modelled |
46.9 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:botulinum neurotoxin type b;
PDBTitle: crystal structure of an engineered botulinum neurotoxin2 type b-derivative, lc-b-gs-hn-b
|
| 68 | d2f22a1 |
|
not modelled |
46.3 |
36 |
Fold:DinB/YfiT-like putative metalloenzymes
Superfamily:DinB/YfiT-like putative metalloenzymes
Family:DinB-like |
| 69 | c1s0bA_ |
|
not modelled |
45.0 |
26 |
PDB header:toxin, hydrolase
Chain: A: PDB Molecule:botulinum neurotoxin type b;
PDBTitle: crystal structure of botulinum neurotoxin type b at ph 4.0
|
| 70 | c3pl4A_ |
|
not modelled |
44.7 |
12 |
PDB header:motor protein
Chain: A: PDB Molecule:flagellar motor switch protein;
PDBTitle: crystal structure of flig (residue 116-343) from h. pylori
|
| 71 | c3zurA_ |
|
not modelled |
43.8 |
13 |
PDB header:hydrolase/signaling protein
Chain: A: PDB Molecule:botulinum neurotoxin type a, synaptosomal-associated
PDBTitle: crystal structure of an engineered botulinum neurotoxin2 type a-snare23 derivative, lc0-a-snap25-hn-a
|
| 72 | c3dl1A_ |
|
not modelled |
39.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metal-dependent hydrolase;
PDBTitle: crystal structure of a putative metal-dependent hydrolase2 (yp_001336084.1) from klebsiella pneumoniae subsp. pneumoniae mgh3 78578 at 2.20 a resolution
|
| 73 | c2y0oA_ |
|
not modelled |
38.9 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:probable d-lyxose ketol-isomerase;
PDBTitle: the structure of a d-lyxose isomerase from the sigmab2 regulon of bacillus subtilis
|
| 74 | c2l0rA_ |
|
not modelled |
38.7 |
15 |
PDB header:hydrolase,toxin
Chain: A: PDB Molecule:lethal factor;
PDBTitle: conformational dynamics of the anthrax lethal factor catalytic center
|
| 75 | d1lmla_ |
|
not modelled |
38.4 |
19 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Leishmanolysin |
| 76 | c3sohB_ |
|
not modelled |
36.4 |
26 |
PDB header:motor protein
Chain: B: PDB Molecule:flagellar motor switch protein flig;
PDBTitle: architecture of the flagellar rotor
|
| 77 | d1bqqm_ |
|
not modelled |
35.5 |
45 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 78 | d2a1jb1 |
|
not modelled |
34.8 |
20 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |
| 79 | d1rm8a_ |
|
not modelled |
32.3 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 80 | c2xs4A_ |
|
not modelled |
32.1 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:karilysin protease;
PDBTitle: structure of karilysin catalytic mmp domain in complex with2 magnesium
|
| 81 | d1hv5a_ |
|
not modelled |
32.0 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 82 | c2x7mA_ |
|
not modelled |
31.3 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:archaemetzincin;
PDBTitle: crystal structure of archaemetzincin (amza) from methanopyrus2 kandleri at 1.5 a resolution
|
| 83 | d1cxva_ |
|
not modelled |
30.2 |
19 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 84 | d1i76a_ |
|
not modelled |
29.7 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 85 | d1hfca_ |
|
not modelled |
28.9 |
45 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 86 | d1mmqa_ |
|
not modelled |
28.5 |
27 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 87 | d1xuca1 |
|
not modelled |
28.5 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 88 | c2k9mA_ |
|
not modelled |
28.2 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpon;
PDBTitle: structure of the core binding domain of sigma54
|
| 89 | d1qiba_ |
|
not modelled |
27.9 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 90 | d1y93a1 |
|
not modelled |
27.9 |
27 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 91 | d1q3aa_ |
|
not modelled |
27.5 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 92 | d1hy7a_ |
|
not modelled |
27.5 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 93 | d2ovxa1 |
|
not modelled |
27.4 |
27 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 94 | d1cgla_ |
|
not modelled |
27.3 |
45 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 95 | d1lsha1 |
|
not modelled |
25.9 |
13 |
Fold:alpha-alpha superhelix
Superfamily:Lipovitellin-phosvitin complex, superhelical domain
Family:Lipovitellin-phosvitin complex, superhelical domain |
| 96 | d1s5qb_ |
|
not modelled |
25.7 |
11 |
Fold:PAH2 domain
Superfamily:PAH2 domain
Family:PAH2 domain |
| 97 | c2py8B_ |
|
not modelled |
25.4 |
20 |
PDB header:chaperone
Chain: B: PDB Molecule:hypothetical protein rbcx;
PDBTitle: rbcx
|
| 98 | c2jsdA_ |
|
not modelled |
25.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:matrix metalloproteinase-20;
PDBTitle: solution structure of mmp20 complexed with nngh
|
| 99 | d1vh8a_ |
|
not modelled |
24.8 |
10 |
Fold:Bacillus chorismate mutase-like
Superfamily:IpsF-like
Family:IpsF-like |
| 100 | d1x2ia1 |
|
not modelled |
24.8 |
15 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |
| 101 | d1hova_ |
|
not modelled |
24.8 |
23 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 102 | d1kfta_ |
|
not modelled |
23.5 |
24 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Excinuclease UvrC C-terminal domain |
| 103 | c1kftA_ |
|
not modelled |
23.5 |
24 |
PDB header:dna binding protein
Chain: A: PDB Molecule:excinuclease abc subunit c;
PDBTitle: solution structure of the c-terminal domain of uvrc from e-2 coli
|
| 104 | d1st6a5 |
|
not modelled |
23.3 |
14 |
Fold:Four-helical up-and-down bundle
Superfamily:alpha-catenin/vinculin-like
Family:alpha-catenin/vinculin |
| 105 | d2py8a1 |
|
not modelled |
23.0 |
22 |
Fold:RbcX-like
Superfamily:RbcX-like
Family:RbcX-like |
| 106 | c2veeC_ |
|
not modelled |
22.6 |
8 |
PDB header:transport protein
Chain: C: PDB Molecule:protoglobin;
PDBTitle: structure of protoglobin from methanosarcina acetivorans2 c2a
|
| 107 | d1eaka2 |
|
not modelled |
22.5 |
27 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 108 | d2vv5a2 |
|
not modelled |
21.9 |
11 |
Fold:Ferredoxin-like
Superfamily:Mechanosensitive channel protein MscS (YggB), C-terminal domain
Family:Mechanosensitive channel protein MscS (YggB), C-terminal domain |
| 109 | c2z0uB_ |
|
not modelled |
21.7 |
10 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:ww domain-containing protein 1;
PDBTitle: crystal structure of c2 domain of kibra protein
|
| 110 | c3pkrA_ |
|
not modelled |
21.7 |
12 |
PDB header:motor protein
Chain: A: PDB Molecule:flagellar motor switch protein;
PDBTitle: crystal structure of flig (residue 86-343) from h. pylori
|
| 111 | d1fbla2 |
|
not modelled |
21.4 |
45 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 112 | d1j9ia_ |
|
not modelled |
20.6 |
22 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:Terminase gpNU1 subunit domain |
| 113 | d1c7ka_ |
|
not modelled |
20.3 |
56 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Zinc protease |
| 114 | c2cltB_ |
|
not modelled |
20.1 |
36 |
PDB header:hydrolase
Chain: B: PDB Molecule:interstitial collagenase;
PDBTitle: crystal structure of the active form (full-length) of human2 fibroblast collagenase.
|