1 c2j0wA_
100.0
100
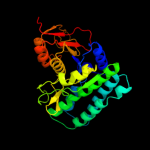
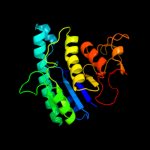
PDB header: transferaseChain: A: PDB Molecule: lysine-sensitive aspartokinase 3;PDBTitle: crystal structure of e. coli aspartokinase iii in complex2 with aspartate and adp (r-state)
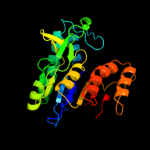
2 c2cdqB_
100.0
34
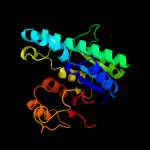
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of arabidopsis thaliana aspartate kinase2 complexed with lysine and s-adenosylmethionine
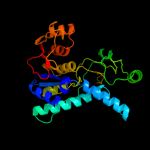
3 c3c1nA_
100.0
30
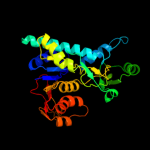
PDB header: transferaseChain: A: PDB Molecule: probable aspartokinase;PDBTitle: crystal structure of allosteric inhibition threonine-sensitive2 aspartokinase from methanococcus jannaschii with l-threonine
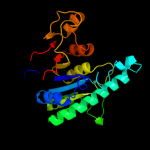
4 c3l76B_
100.0
30
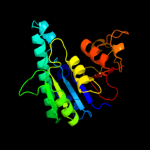
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of aspartate kinase from synechocystis
5 c3ab4K_
100.0
27
PDB header: transferaseChain: K: PDB Molecule: aspartokinase;PDBTitle: crystal structure of feedback inhibition resistant mutant of aspartate2 kinase from corynebacterium glutamicum in complex with lysine and3 threonine
6 d2cdqa1
100.0
33
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like7 d2j0wa1
100.0
100
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like8 d2hmfa1
100.0
35
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like9 d1ybda1
100.0
21
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like10 c2e9yA_
100.0
20
PDB header: transferaseChain: A: PDB Molecule: carbamate kinase;PDBTitle: crystal structure of project ape1968 from aeropyrum pernix k1
11 d2bnea1
100.0
25
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like12 c3k4yB_
100.0
17
PDB header: transferaseChain: B: PDB Molecule: isopentenyl phosphate kinase;PDBTitle: crystal structure of isopentenyl phosphate kinase from m. jannaschii2 in complex with ipp
13 c3ll9A_
100.0
20
PDB header: transferaseChain: A: PDB Molecule: isopentenyl phosphate kinase;PDBTitle: x-ray structures of isopentenyl phosphate kinase
14 c3ek5A_
100.0
25
PDB header: transferaseChain: A: PDB Molecule: uridylate kinase;PDBTitle: unique gtp-binding pocket and allostery of ump kinase from a gram-2 negative phytopathogen bacterium
15 d2a1fa1
100.0
24
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like16 d1z9da1
100.0
24
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like17 d1gs5a_
100.0
19
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase18 d1e19a_
99.9
15
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: Carbamate kinase19 c3nwyB_
99.9
25
PDB header: transferaseChain: B: PDB Molecule: uridylate kinase;PDBTitle: structure and allosteric regulation of the uridine monophosphate2 kinase from mycobacterium tuberculosis
20 d2bufa1
99.9
20
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase21 c2jjxC_
not modelled
99.9
20
PDB header: transferaseChain: C: PDB Molecule: uridylate kinase;PDBTitle: the crystal structure of ump kinase from bacillus anthracis2 (ba1797)
22 c3l86A_
not modelled
99.9
19
PDB header: transferaseChain: A: PDB Molecule: acetylglutamate kinase;PDBTitle: the crystal structure of smu.665 from streptococcus mutans ua159
23 c2rd5A_
not modelled
99.9
18
PDB header: protein bindingChain: A: PDB Molecule: acetylglutamate kinase-like protein;PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
24 d2brxa1
not modelled
99.9
26
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like25 d1b7ba_
not modelled
99.9
16
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: Carbamate kinase26 c3ll5C_
not modelled
99.9
18
PDB header: transferaseChain: C: PDB Molecule: gamma-glutamyl kinase related protein;PDBTitle: crystal structure of t. acidophilum isopentenyl phosphate kinase2 product complex
27 d2ij9a1
not modelled
99.9
25
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like28 c2zhoB_
not modelled
99.9
20
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of the regulatory subunit of aspartate2 kinase from thermus thermophilus (ligand free form)
29 c2egxA_
not modelled
99.9
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative acetylglutamate kinase;PDBTitle: crystal structure of the putative acetylglutamate kinase from thermus2 thermophilus
30 c2va1A_
not modelled
99.9
23
PDB header: transferaseChain: A: PDB Molecule: uridylate kinase;PDBTitle: crystal structure of ump kinase from ureaplasma parvum
31 c2dtjA_
not modelled
99.9
19
PDB header: transferaseChain: A: PDB Molecule: aspartokinase;PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
32 c3mahA_
not modelled
99.9
29
PDB header: transferaseChain: A: PDB Molecule: aspartokinase;PDBTitle: a putative c-terminal regulatory domain of aspartate kinase from2 porphyromonas gingivalis w83.
33 d2btya1
not modelled
99.9
18
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase34 c2re1A_
not modelled
99.9
18
PDB header: transferaseChain: A: PDB Molecule: aspartokinase, alpha and beta subunits;PDBTitle: crystal structure of aspartokinase alpha and beta subunits
35 d2akoa1
not modelled
99.9
17
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like36 c2j4kC_
not modelled
99.9
23
PDB header: transferaseChain: C: PDB Molecule: uridylate kinase;PDBTitle: crystal structure of uridylate kinase from sulfolobus2 solfataricus in complex with ump to 2.2 angstrom3 resolution
37 c2v5hB_
not modelled
99.9
17
PDB header: transcriptionChain: B: PDB Molecule: acetylglutamate kinase;PDBTitle: controlling the storage of nitrogen as arginine: the2 complex of pii and acetylglutamate kinase from3 synechococcus elongatus pcc 7942
38 c2w21A_
not modelled
99.9
22
PDB header: transferaseChain: A: PDB Molecule: glutamate 5-kinase;PDBTitle: crystal structure of the aminoacid kinase domain of the2 glutamate 5 kinase of escherichia coli.
39 c3d40A_
not modelled
99.9
22
PDB header: transferaseChain: A: PDB Molecule: foma protein;PDBTitle: crystal structure of fosfomycin resistance kinase foma from2 streptomyces wedmorensis complexed with diphosphate
40 d2ap9a1
not modelled
99.9
24
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase41 c2j5tF_
not modelled
99.9
21
PDB header: transferaseChain: F: PDB Molecule: glutamate 5-kinase;PDBTitle: glutamate 5-kinase from escherichia coli complexed with2 glutamate
42 c2ogxB_
not modelled
99.8
24
PDB header: metal binding proteinChain: B: PDB Molecule: molybdenum storage protein subunit beta;PDBTitle: the crystal structure of the molybdenum storage protein from2 azotobacter vinelandii loaded with polyoxotungstates (wsto)
43 c3kzfC_
not modelled
99.8
19
PDB header: transferaseChain: C: PDB Molecule: carbamate kinase;PDBTitle: structure of giardia carbamate kinase
44 c2ogxA_
not modelled
99.8
24
PDB header: metal binding proteinChain: A: PDB Molecule: molybdenum storage protein subunit alpha;PDBTitle: the crystal structure of the molybdenum storage protein from2 azotobacter vinelandii loaded with polyoxotungstates (wsto)
45 c2f06B_
not modelled
99.6
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
46 c2r98A_
not modelled
99.6
18
PDB header: transferaseChain: A: PDB Molecule: putative acetylglutamate synthase;PDBTitle: crystal structure of n-acetylglutamate synthase (selenomet2 substituted) from neisseria gonorrhoeae
47 d2j0wa3
not modelled
99.5
100
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like48 d2hmfa2
not modelled
99.4
28
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like49 d2cdqa3
not modelled
99.4
40
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like50 d2hmfa3
not modelled
99.3
21
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like51 d2cdqa2
not modelled
99.2
37
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like52 d2j0wa2
not modelled
99.1
100
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like53 c2nyiB_
not modelled
96.5
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: unknown protein;PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
54 c1u8sB_
not modelled
95.9
14
PDB header: transcriptionChain: B: PDB Molecule: glycine cleavage system transcriptionalPDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
55 c1zhvA_
not modelled
93.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein atu0741;PDBTitle: x-ray crystal structure protein atu0741 from agobacterium tumefaciens.2 northeast structural genomics consortium target atr8.
56 d1zhva2
not modelled
92.3
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Atu0741-like57 c2fgcA_
not modelled
91.5
19
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase, small subunit;PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
58 c2pc6C_
not modelled
89.8
13
PDB header: lyaseChain: C: PDB Molecule: probable acetolactate synthase isozyme iii (small subunit);PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
59 c2f1fA_
not modelled
88.5
14
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase isozyme iii small subunit;PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
60 d2f06a1
not modelled
88.5
10
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like61 d1zvpa2
not modelled
87.5
21
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: VC0802-like62 c1zvpB_
not modelled
87.3
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein vc0802;PDBTitle: crystal structure of a protein of unknown function vc0802 from vibrio2 cholerae, possible transport protein
63 d2pc6a2
not modelled
83.9
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like64 d2fgca2
not modelled
83.7
17
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like65 d2f1fa1
not modelled
82.7
12
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like66 d1ygya3
not modelled
81.5
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain67 d2f06a2
not modelled
78.3
29
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like68 d1sc6a3
not modelled
77.3
11
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain69 d1u8sa2
not modelled
76.3
10
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor70 c3nrbD_
not modelled
76.3
16
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
71 c2qmxB_
not modelled
75.9
13
PDB header: ligaseChain: B: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
72 d1zpva1
not modelled
75.4
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: SP0238-like73 c3ibwA_
not modelled
74.1
17
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
74 d1phza1
not modelled
70.8
17
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain75 d1u8sa1
not modelled
70.3
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor76 c3mwbA_
not modelled
68.8
16
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
77 c1ygyA_
not modelled
64.9
8
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
78 d2qmwa2
not modelled
62.5
19
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain79 c2qmwA_
not modelled
58.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of the prephenate dehydratase (pdt) from2 staphylococcus aureus subsp. aureus mu50
80 c3luyA_
not modelled
56.1
7
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
81 c2vcbA_
not modelled
44.9
16
PDB header: hydrolaseChain: A: PDB Molecule: alpha-n-acetylglucosaminidase;PDBTitle: family 89 glycoside hydrolase from clostridium perfringens2 in complex with pugnac
82 d1a9xa4
not modelled
42.4
15
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like83 c2k4mA_
not modelled
40.8
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0146 protein mth_1000;PDBTitle: solution nmr structure of m. thermoautotrophicum protein2 mth_1000, northeast structural genomics consortium target3 tr8
84 c3n0vD_
not modelled
40.5
12
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
85 c2nu9E_
not modelled
39.6
31
PDB header: ligaseChain: E: PDB Molecule: succinyl-coa synthetase beta chain;PDBTitle: c123at mutant of e. coli succinyl-coa synthetase2 orthorhombic crystal form
86 d2g0ta1
not modelled
38.4
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like87 c3cuxA_
not modelled
35.1
21
PDB header: transferaseChain: A: PDB Molecule: malate synthase;PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
88 c3o1lB_
not modelled
35.0
14
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
89 c3mtjA_
not modelled
34.7
25
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
90 c2fvgA_
not modelled
33.3
11
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: crystal structure of endoglucanase (tm1049) from thermotoga maritima2 at 2.01 a resolution
91 d1tzoa_
not modelled
33.3
23
Fold: Anthrax protective antigenSuperfamily: Anthrax protective antigenFamily: Anthrax protective antigen92 c3nbuC_
not modelled
27.0
21
PDB header: isomeraseChain: C: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of pgi glucosephosphate isomerase
93 c3cuzA_
not modelled
25.2
20
PDB header: transferaseChain: A: PDB Molecule: malate synthase a;PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
94 c2wu8A_
not modelled
24.4
17
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: structural studies of phosphoglucose isomerase from2 mycobacterium tuberculosis h37rv
95 d2fvga1
not modelled
24.2
23
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain96 c3ewiB_
not modelled
24.1
21
PDB header: transferaseChain: B: PDB Molecule: n-acylneuraminate cytidylyltransferase;PDBTitle: structural analysis of the c-terminal domain of murine cmp-2 sialic acid synthetase
97 c3isrB_
not modelled
22.1
20
PDB header: hydrolaseChain: B: PDB Molecule: transglutaminase-like enzymes, putative cysteine protease;PDBTitle: the crystal structure of a putative cysteine protease from cytophaga2 hutchinsonii to 1.9a
98 d1gzda_
not modelled
22.0
23
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI99 c3ljkA_
not modelled
21.4
17
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: glucose-6-phosphate isomerase from francisella tularensis.
100 c2o2cB_
not modelled
21.2
19
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase, glycosomal;PDBTitle: crystal structure of phosphoglucose isomerase from t. brucei2 containing glucose-6-phosphate in the active site
101 c3l7pA_
not modelled
20.8
6
PDB header: transcriptionChain: A: PDB Molecule: putative nitrogen regulatory protein pii;PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
102 c3hjbA_
not modelled
20.7
23
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: 1.5 angstrom crystal structure of glucose-6-phosphate isomerase from2 vibrio cholerae.