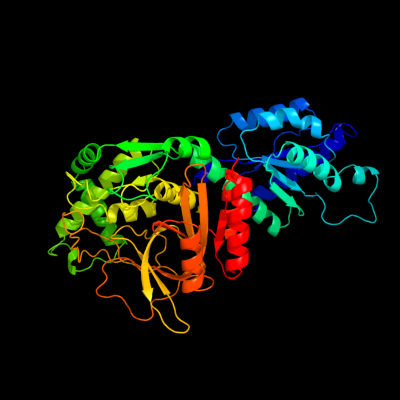
| 1 | c2hxgB_
|
|
 |
100.0 |
99 |
PDB header:isomerase
Chain: B: PDB Molecule:l-arabinose isomerase;
PDBTitle: crystal structure of mn2+ bound ecai
|
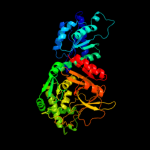
| 2 | d2ajta2
|
|
 |
100.0 |
99 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:AraA N-terminal and middle domain-like |
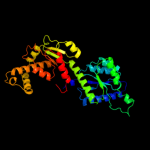
| 3 | d2ajta1
|
|
 |
100.0 |
99 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:FucI/AraA C-terminal domain-like
Family:AraA C-terminal domain-like |
| 4 | d1fuia2
|
|
 |
100.0 |
12 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:L-fucose isomerase, N-terminal and second domains |
| 5 | c1fuiB_
|
|
 |
100.0 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:l-fucose isomerase;
PDBTitle: l-fucose isomerase from escherichia coli
|
| 6 | c3a9rA_
|
|
 |
100.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:d-arabinose isomerase;
PDBTitle: x-ray structures of bacillus pallidus d-arabinose2 isomerasecomplex with (4r)-2-methylpentane-2,4-diol
|
| 7 | c2c2xB_
|
|
 |
87.2 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase-
PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
|
| 8 | d1a4ia2
|
|
 |
86.5 |
26 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Tetrahydrofolate dehydrogenase/cyclohydrolase |
| 9 | d1b0aa2
|
|
 |
84.7 |
23 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Tetrahydrofolate dehydrogenase/cyclohydrolase |
| 10 | c3p2oA_
|
|
 |
82.3 |
22 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 11 | c3nglA_
|
|
 |
81.6 |
15 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
|
| 12 | c3p2oB_
|
|
 |
79.3 |
21 |
PDB header:oxidoreductase, hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 13 | c1a4iB_
|
|
 |
77.9 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
| 14 | c1b0aA_
|
|
 |
74.8 |
23 |
PDB header:oxidoreductase,hydrolase
Chain: A: PDB Molecule:protein (fold bifunctional protein);
PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
|
| 15 | d1gcaa_
|
|
 |
74.7 |
12 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 16 | d1fuia1
|
|
 |
68.2 |
15 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:FucI/AraA C-terminal domain-like
Family:L-fucose isomerase, C-terminal domain |
| 17 | c4a5oB_
|
|
 |
67.3 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of pseudomonas aeruginosa n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
| 18 | c3h5oB_
|
|
 |
65.2 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator gntr;
PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
|
| 19 | c4a26B_
|
|
 |
64.3 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative c-1-tetrahydrofolate synthase, cytoplasmic;
PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
|
| 20 | c2p2gD_
|
|
 |
64.1 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from mycobacterium2 tuberculosis (rv1656): orthorhombic form
|
| 21 | c3l07B_ |
|
not modelled |
63.4 |
17 |
PDB header:oxidoreductase,hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
|
| 22 | c2x7xA_ |
|
not modelled |
61.2 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: fructose binding periplasmic domain of hybrid two component2 system bt1754
|
| 23 | c3bblA_ |
|
not modelled |
60.5 |
12 |
PDB header:regulatory protein
Chain: A: PDB Molecule:regulatory protein of laci family;
PDBTitle: crystal structure of a regulatory protein of laci family from2 chloroflexus aggregans
|
| 24 | c3oqbF_ |
|
not modelled |
57.9 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from bradyrhizobium2 japonicum usda 110
|
| 25 | c3v5nA_ |
|
not modelled |
56.2 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of oxidoreductase from sinorhizobium meliloti
|
| 26 | c3nt5B_ |
|
not modelled |
56.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
| 27 | c3eafA_ |
|
not modelled |
54.3 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter, substrate binding protein;
PDBTitle: crystal structure of abc transporter, substrate binding protein2 aeropyrum pernix
|
| 28 | d2dria_ |
|
not modelled |
53.8 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 29 | c3kkeA_ |
|
not modelled |
51.4 |
12 |
PDB header:transcription regulator
Chain: A: PDB Molecule:laci family transcriptional regulator;
PDBTitle: crystal structure of a laci family transcriptional regulator2 from mycobacterium smegmatis
|
| 30 | c3e18A_ |
|
not modelled |
51.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 31 | c3ec7C_ |
|
not modelled |
51.2 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase;
PDBTitle: crystal structure of putative dehydrogenase from salmonella2 typhimurium lt2
|
| 32 | d1skyb3 |
|
not modelled |
50.2 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 33 | c3ksmA_ |
|
not modelled |
47.1 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
| 34 | d2fvya1 |
|
not modelled |
46.5 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 35 | c3d8uA_ |
|
not modelled |
45.6 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:purr transcriptional regulator;
PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
|
| 36 | c3moiA_ |
|
not modelled |
44.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
| 37 | c2xecD_ |
|
not modelled |
43.7 |
14 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 38 | c3hcwB_ |
|
not modelled |
43.0 |
8 |
PDB header:rna binding protein
Chain: B: PDB Molecule:maltose operon transcriptional repressor;
PDBTitle: crystal structure of probable maltose operon transcriptional repressor2 malr from staphylococcus areus
|
| 39 | c3ctpB_ |
|
not modelled |
41.9 |
11 |
PDB header:transcription regulator
Chain: B: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of periplasmic binding protein/laci transcriptional2 regulator from alkaliphilus metalliredigens qymf complexed with d-3 xylulofuranose
|
| 40 | c3gfgB_ |
|
not modelled |
41.2 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized oxidoreductase yvaa;
PDBTitle: structure of putative oxidoreductase yvaa from bacillus subtilis in2 triclinic form
|
| 41 | d8abpa_ |
|
not modelled |
41.0 |
12 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 42 | d1edza2 |
|
not modelled |
40.6 |
27 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Tetrahydrofolate dehydrogenase/cyclohydrolase |
| 43 | d1hcza2 |
|
not modelled |
40.3 |
28 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
| 44 | c3l49D_ |
|
not modelled |
39.5 |
9 |
PDB header:transport protein
Chain: D: PDB Molecule:abc sugar (ribose) transporter, periplasmic
PDBTitle: crystal structure of abc sugar transporter subunit from2 rhodobacter sphaeroides 2.4.1
|
| 45 | d1dxha2 |
|
not modelled |
39.2 |
14 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 46 | c3kuxA_ |
|
not modelled |
39.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the ypo2259 putative oxidoreductase from yersinia pestis
|
| 47 | c1h6dL_ |
|
not modelled |
39.0 |
13 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 48 | d2jdia3 |
|
not modelled |
38.3 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 49 | c3gr7A_ |
|
not modelled |
38.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 50 | d1gg2g_ |
|
not modelled |
37.8 |
20 |
Fold:Non-globular all-alpha subunits of globular proteins
Superfamily:Transducin (heterotrimeric G protein), gamma chain
Family:Transducin (heterotrimeric G protein), gamma chain |
| 51 | c2r9vA_ |
|
not modelled |
37.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp synthase subunit alpha;
PDBTitle: crystal structure of atp synthase subunit alpha (tm1612) from2 thermotoga maritima at 2.10 a resolution
|
| 52 | c3c1aB_ |
|
not modelled |
37.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase (zp_00056571.1) from2 magnetospirillum magnetotacticum ms-1 at 1.85 a resolution
|
| 53 | d2b7oa1 |
|
not modelled |
37.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class-II DAHP synthetase |
| 54 | c3ceaA_ |
|
not modelled |
35.0 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 55 | d1omwg_ |
|
not modelled |
34.9 |
20 |
Fold:Non-globular all-alpha subunits of globular proteins
Superfamily:Transducin (heterotrimeric G protein), gamma chain
Family:Transducin (heterotrimeric G protein), gamma chain |
| 56 | c2qu7B_ |
|
not modelled |
34.4 |
5 |
PDB header:transcription
Chain: B: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of a putative transcription regulator2 from staphylococcus saprophyticus subsp. saprophyticus
|
| 57 | c3oa2B_ |
|
not modelled |
34.3 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from pseudomonas2 aeruginosa in complex with nad at 1.5 angstrom resolution
|
| 58 | c1ofgF_ |
|
not modelled |
34.2 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 59 | d1lc0a1 |
|
not modelled |
34.0 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 60 | d1gqoa_ |
|
not modelled |
33.7 |
15 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 61 | d2nu7b1 |
|
not modelled |
33.6 |
13 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 62 | c3qk7C_ |
|
not modelled |
32.9 |
15 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcriptional regulator from yersinia2 pestis biovar microtus str. 91001
|
| 63 | c1edzA_ |
|
not modelled |
32.6 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5,10-methylenetetrahydrofolate dehydrogenase;
PDBTitle: structure of the nad-dependent 5,10-2 methylenetetrahydrofolate dehydrogenase from saccharomyces3 cerevisiae
|
| 64 | d2gm3a1 |
|
not modelled |
32.5 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 65 | c1kxfA_ |
|
not modelled |
32.2 |
26 |
PDB header:viral protein
Chain: A: PDB Molecule:sindbis virus capsid protein;
PDBTitle: sindbis virus capsid, (wild-type) residues 1-264,2 tetragonal crystal form (form ii)
|
| 66 | d1auoa_ |
|
not modelled |
32.1 |
13 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase/thioesterase 1 |
| 67 | c3fd8A_ |
|
not modelled |
31.8 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 68 | c2rjoA_ |
|
not modelled |
31.7 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:twin-arginine translocation pathway signal protein;
PDBTitle: crystal structure of twin-arginine translocation pathway signal2 protein from burkholderia phytofirmans
|
| 69 | d1ep5a_ |
|
not modelled |
31.5 |
28 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
| 70 | c3lkbB_ |
|
not modelled |
31.4 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:probable branched-chain amino acid abc
PDBTitle: crystal structure of a branched chain amino acid abc2 transporter from thermus thermophilus with bound valine
|
| 71 | c1zq1B_ |
|
not modelled |
31.1 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
|
| 72 | c3oa0B_ |
|
not modelled |
30.8 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lipopolysaccaride biosynthesis protein wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from thermus2 thermophilus in complex with nad and udp-glcnaca
|
| 73 | c3cn9B_ |
|
not modelled |
30.7 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:carboxylesterase;
PDBTitle: crystal structure analysis of the carboxylesterase pa3859 from2 pseudomonas aeruginosa pao1- orthorhombic crystal form
|
| 74 | c1zh8B_ |
|
not modelled |
30.7 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 75 | d1wyka_ |
|
not modelled |
30.5 |
26 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
| 76 | d1ci3m2 |
|
not modelled |
30.5 |
21 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
| 77 | c1tltB_ |
|
not modelled |
29.3 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase (virulence factor mvim homolog);
PDBTitle: crystal structure of a putative oxidoreductase (virulence factor mvim2 homolog)
|
| 78 | c3f4lF_ |
|
not modelled |
29.3 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative oxidoreductase yhhx;
PDBTitle: crystal structure of a probable oxidoreductase yhhx in2 triclinic form. northeast structural genomics target er647
|
| 79 | d1e2wa2 |
|
not modelled |
29.2 |
32 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
| 80 | d1fx0a3 |
|
not modelled |
28.7 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 81 | c2uygF_ |
|
not modelled |
28.4 |
13 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
|
| 82 | c3bdkB_ |
|
not modelled |
28.2 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
| 83 | c2glxD_ |
|
not modelled |
28.0 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
| 84 | d1gtza_ |
|
not modelled |
27.7 |
19 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 85 | d1ekxa2 |
|
not modelled |
27.4 |
20 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 86 | c3g85A_ |
|
not modelled |
27.3 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator (laci family);
PDBTitle: crystal structure of laci family transcription regulator from2 clostridium acetobutylicum
|
| 87 | d1h6da1 |
|
not modelled |
27.1 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 88 | c3ezyB_ |
|
not modelled |
27.0 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from2 thermotoga maritima
|
| 89 | d1ydwa1 |
|
not modelled |
26.8 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 90 | c2pfsA_ |
|
not modelled |
26.7 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
|
| 91 | d2g50a2 |
|
not modelled |
26.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 92 | d1xima_ |
|
not modelled |
26.3 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 93 | c2vc6A_ |
|
not modelled |
26.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 94 | c2p10D_ |
|
not modelled |
26.0 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 95 | c3sdoB_ |
|
not modelled |
25.9 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
| 96 | d2c4va1 |
|
not modelled |
25.2 |
19 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 97 | d2h1ia1 |
|
not modelled |
25.2 |
13 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase/thioesterase 1 |
| 98 | c1jyeA_ |
|
not modelled |
25.0 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:lactose operon repressor;
PDBTitle: structure of a dimeric lac repressor with c-terminal deletion and k84l2 substitution
|
| 99 | d1jyea_ |
|
not modelled |
25.0 |
15 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 100 | c1xdwA_ |
|
not modelled |
24.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad+-dependent (r)-2-hydroxyglutarate
PDBTitle: nad+-dependent (r)-2-hydroxyglutarate dehydrogenase from2 acidaminococcus fermentans
|
| 101 | c1lc3A_ |
|
not modelled |
24.7 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biliverdin reductase a;
PDBTitle: crystal structure of a biliverdin reductase enzyme-cofactor2 complex
|
| 102 | d1otha2 |
|
not modelled |
24.7 |
18 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 103 | d2glka1 |
|
not modelled |
24.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 104 | c3pf6C_ |
|
not modelled |
24.5 |
39 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein pp-luz7_gp033;
PDBTitle: the structure of uncharacterized protein pp-luz7_gp033 from2 pseudomonas phage luz7.
|
| 105 | c3gv0A_ |
|
not modelled |
24.3 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
|
| 106 | c3ip3D_ |
|
not modelled |
24.3 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, putative;
PDBTitle: structure of putative oxidoreductase (tm_0425) from2 thermotoga maritima
|
| 107 | c1kmhA_ |
|
not modelled |
24.1 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:atpase alpha subunit;
PDBTitle: crystal structure of spinach chloroplast f1-atpase2 complexed with tentoxin
|
| 108 | c2ixaA_ |
|
not modelled |
23.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 109 | c2zkqj_ |
|
not modelled |
23.9 |
18 |
PDB header:ribosomal protein/rna
Chain: J: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 110 | c3db2C_ |
|
not modelled |
23.8 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 111 | d2a9da2 |
|
not modelled |
23.7 |
20 |
Fold:Oxidoreductase molybdopterin-binding domain
Superfamily:Oxidoreductase molybdopterin-binding domain
Family:Oxidoreductase molybdopterin-binding domain |
| 112 | c2ehhE_ |
|
not modelled |
23.3 |
13 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 113 | d1y81a1 |
|
not modelled |
23.0 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 114 | d1uc2a_ |
|
not modelled |
22.8 |
26 |
Fold:Hypothetical protein PH1602
Superfamily:Hypothetical protein PH1602
Family:Hypothetical protein PH1602 |
| 115 | d1ryda1 |
|
not modelled |
22.7 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 116 | c2o48X_ |
|
not modelled |
22.6 |
18 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 117 | c3bi8A_ |
|
not modelled |
22.3 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 118 | c3eb2A_ |
|
not modelled |
22.2 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 119 | d1jx6a_ |
|
not modelled |
22.2 |
10 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 120 | c2qvcC_ |
|
not modelled |
22.1 |
12 |
PDB header:transport protein
Chain: C: PDB Molecule:sugar abc transporter, periplasmic sugar-binding
PDBTitle: crystal structure of a periplasmic sugar abc transporter2 from thermotoga maritima
|