1 c2dskA_
100.0
34
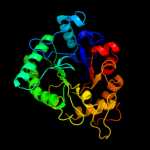
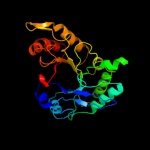
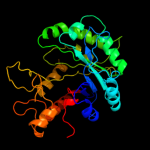
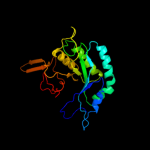
PDB header: hydrolaseChain: A: PDB Molecule: chitinase;PDBTitle: crystal structure of catalytic domain of hyperthermophilic chitinase2 from pyrococcus furiosus
2 c3ianA_
100.0
16
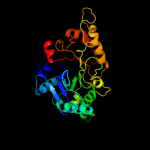
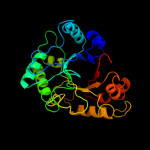
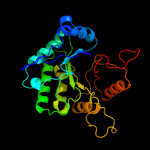
PDB header: hydrolaseChain: A: PDB Molecule: chitinase;PDBTitle: crystal structure of a chitinase from lactococcus lactis2 subsp. lactis
3 c3ebvA_
99.9
15
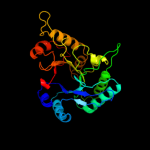
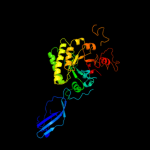
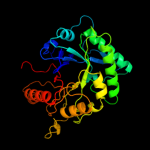
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: chinitase a;PDBTitle: crystal structure of putative chitinase a from streptomyces2 coelicolor.
4 d2hvma_
99.9
19
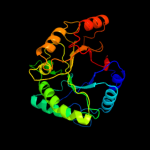
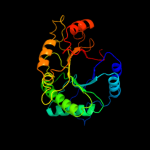
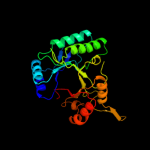
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase5 c3d5hA_
99.9
14
PDB header: protein bindingChain: A: PDB Molecule: haementhin;PDBTitle: crystal structure of haementhin from haemanthus multiflorus2 at 2.0a resolution: formation of a novel loop on a tim3 barrel fold and its functional significance
6 c2uy2A_
99.8
18
PDB header: hydrolaseChain: A: PDB Molecule: endochitinase;PDBTitle: sccts1_apo crystal structure
7 c3n12A_
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: chitinase a;PDBTitle: crystal stricture of chitinase in complex with zinc atoms from2 bacillus cereus nctu2
8 c2gsjA_
99.8
18
PDB header: hydrolaseChain: A: PDB Molecule: protein ppl-2;PDBTitle: cdna cloning and 1.75a crystal structure determination of2 ppl2, a novel chimerolectin from parkia platycephala seeds3 exhibiting endochitinolytic activity
9 d1cnva_
99.8
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase10 c3b9eA_
99.7
20
PDB header: hydrolaseChain: A: PDB Molecule: chitinase a;PDBTitle: crystal structure of inactive mutant e315m chitinase a from2 vibrio harveyi
11 c2xucA_
99.7
13
PDB header: hydrolaseChain: A: PDB Molecule: chitinase;PDBTitle: natural product-guided discovery of a fungal chitinase2 inhibitor
12 d1itxa1
99.7
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase13 c1ur8B_
99.7
20
PDB header: hydrolaseChain: B: PDB Molecule: chitinase b;PDBTitle: interactions of a family 18 chitinase with the designed2 inhibitor hm508, and its degradation product,3 chitobiono-delta-lactone
14 c1rd6A_
99.7
14
PDB header: hydrolaseChain: A: PDB Molecule: chitinase a;PDBTitle: crystal structure of s. marcescens chitinase a mutant w167a
15 d1edqa2
99.6
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase16 c3co4A_
99.6
11
PDB header: hydrolaseChain: A: PDB Molecule: chitinase;PDBTitle: crystal structure of a chitinase from bacteroides2 thetaiotaomicron
17 d1jnda1
99.6
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase18 d1w9pa1
99.6
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase19 d1ta3a_
99.6
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase20 c3pohA_
99.5
14
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-n-acetylglucosaminidase f1;PDBTitle: crystal structure of an endo-beta-n-acetylglucosaminidase (bt_3987)2 from bacteroides thetaiotaomicron vpi-5482 at 1.55 a resolution
21 c1wnoB_
not modelled
99.5
18
PDB header: hydrolaseChain: B: PDB Molecule: chitinase;PDBTitle: crystal structure of a native chitinase from aspergillus fumigatus yj-2 407
22 c1itxA_
not modelled
99.5
19
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase;PDBTitle: catalytic domain of chitinase a1 from bacillus circulans wl-12
23 d1goia2
not modelled
99.5
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase24 d1wb0a1
not modelled
99.5
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase25 d2pi6a1
not modelled
99.5
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase26 c3simA_
not modelled
99.5
15
PDB header: hydrolaseChain: A: PDB Molecule: protein, family 18 chitinase;PDBTitle: crystallographic structure analysis of family 18 chitinase from crocus2 vernus
27 d1hjxa1
not modelled
99.5
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase28 c1ll4A_
not modelled
99.5
20
PDB header: hydrolaseChain: A: PDB Molecule: chitinase 1;PDBTitle: structure of c. immitis chitinase 1 complexed with2 allosamidin
29 d1ll7a1
not modelled
99.5
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase30 c1lq0A_
not modelled
99.4
15
PDB header: hydrolaseChain: A: PDB Molecule: chitotriosidase;PDBTitle: crystal structure of human chitotriosidase at 2.2 angstrom2 resolution
31 c1kfwA_
not modelled
99.4
19
PDB header: hydrolaseChain: A: PDB Molecule: chitinase b;PDBTitle: structure of catalytic domain of psychrophilic chitinase b from2 arthrobacter tad20
32 c3chfA_
not modelled
99.4
15
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: chitinase;PDBTitle: crystal structure of aspergillus fumigatus chitinase b1 in complex2 with tetrapeptide
33 c1hjvA_
not modelled
99.4
15
PDB header: lectinChain: A: PDB Molecule: chitinase-3 like protein 1;PDBTitle: crystal structure of hcgp-39 in complex with chitin2 tetramer
34 d1kfwa1
not modelled
99.4
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase35 c1wb0A_
not modelled
99.4
15
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: chitotriosidase 1;PDBTitle: specificity and affinity of natural product cyclopentapeptide2 inhibitor argifin against human chitinase
36 c3qokA_
not modelled
99.4
18
PDB header: hydrolaseChain: A: PDB Molecule: putative chitinase ii;PDBTitle: crystal structure of putative chitinase ii from klebsiella pneumoniae
37 c3alfA_
not modelled
99.4
15
PDB header: hydrolaseChain: A: PDB Molecule: chitinase, class v;PDBTitle: crystal structure of class v chitinase from nicotiana tobaccum
38 c1jneA_
not modelled
99.4
17
PDB header: hormone/growth factorChain: A: PDB Molecule: imaginal disc growth factor-2;PDBTitle: crystal structure of imaginal disc growth factor-2
39 c3fxyA_
not modelled
99.4
17
PDB header: hydrolaseChain: A: PDB Molecule: acidic mammalian chitinase;PDBTitle: acidic mammalian chinase, catalytic domain
40 d1vf8a1
not modelled
99.4
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase41 c3g6lA_
not modelled
99.4
19
PDB header: hydrolaseChain: A: PDB Molecule: chitinase;PDBTitle: the crystal structure of a chitinase crchi1 from the nematophagous2 fungus clonostachys rosea
42 c4a5qC_
not modelled
99.3
21
PDB header: hydrolaseChain: C: PDB Molecule: chi1;PDBTitle: crystal structure of the chitinase chi1 fitted into the 3d structure2 of the yersinia entomophaga toxin complex
43 c3oa5A_
not modelled
99.3
21
PDB header: hydrolaseChain: A: PDB Molecule: chi1;PDBTitle: the structure of chi1, a chitinase from yersinia entomophaga
44 d1nara_
not modelled
99.3
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase45 c3aquD_
not modelled
99.3
15
PDB header: hydrolaseChain: D: PDB Molecule: at4g19810;PDBTitle: crystal structure of a class v chitinase from arabidopsis thaliana
46 c3bxwB_
not modelled
99.3
12
PDB header: hydrolaseChain: B: PDB Molecule: chitinase domain-containing protein 1;PDBTitle: crystal structure of stabilin-1 interacting chitinase-like2 protein, si-clp
47 c2d49A_
99.2
40
PDB header: hydrolaseChain: A: PDB Molecule: chitinase c;PDBTitle: solution structure of the chitin-binding domain of2 streptomyces griseus chitinase c
48 d1edta_
not modelled
99.2
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase49 c1e9lA_
not modelled
99.2
18
PDB header: macrophage secretory proteinChain: A: PDB Molecule: ym1 secretory protein;PDBTitle: the crystal structure of novel mammalian lectin ym12 suggests a saccharide binding site
50 d2ebna_
not modelled
99.1
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase51 c3cz8A_
not modelled
99.0
15
PDB header: hydrolaseChain: A: PDB Molecule: putative sporulation-specific glycosylase ydhd;PDBTitle: crystal structure of putative sporulation-specific glycosylase ydhd2 from bacillus subtilis
52 d1aiwa_
not modelled
99.0
28
Fold: WW domain-likeSuperfamily: Carbohydrate binding domainFamily: Carbohydrate binding domain53 d1goia1
not modelled
99.0
22
Fold: WW domain-likeSuperfamily: Carbohydrate binding domainFamily: Carbohydrate binding domain54 d1eoka_
not modelled
98.4
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase55 c3b0vD_
not modelled
94.6
13
PDB header: oxidoreductase/rnaChain: D: PDB Molecule: trna-dihydrouridine synthase;PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
56 d1ur4a_
not modelled
94.6
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases57 d2fiqa1
not modelled
90.4
12
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: GatZ-like58 d1ed7a_
not modelled
90.1
35
Fold: WW domain-likeSuperfamily: Carbohydrate binding domainFamily: Carbohydrate binding domain59 c3mi6A_
not modelled
82.6
21
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of the alpha-galactosidase from lactobacillus2 brevis, northeast structural genomics consortium target lbr11.
60 d1nq6a_
not modelled
81.2
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases61 c2wwcA_
not modelled
80.2
14
PDB header: hydrolaseChain: A: PDB Molecule: 1,4-beta-n-acetylmuramidase;PDBTitle: 3d-structure of the modular autolysin lytc from2 streptococcus pneumoniae in complex with synthetic3 peptidoglycan ligand
62 c2aamA_
not modelled
78.4
18
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein tm1410;PDBTitle: crystal structure of a putative glycosidase (tm1410) from thermotoga2 maritima at 2.20 a resolution
63 d2aama1
not modelled
78.4
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: TM1410-like64 d1i1wa_
not modelled
77.3
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases65 c2yfnA_
not modelled
76.5
25
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase-sucrose kinase agask;PDBTitle: galactosidase domain of alpha-galactosidase-sucrose kinase,2 agask
66 c3canA_
not modelled
76.0
13
PDB header: lyase activatorChain: A: PDB Molecule: pyruvate-formate lyase-activating enzyme;PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
67 c2i5gB_
not modelled
74.8
15
PDB header: hydrolaseChain: B: PDB Molecule: amidohydrolase;PDBTitle: crystal strcuture of amidohydrolase from pseudomonas2 aeruginosa
68 c3lu2B_
not modelled
72.7
14
PDB header: hydrolaseChain: B: PDB Molecule: lmo2462 protein;PDBTitle: structure of lmo2462, a listeria monocytogenes amidohydrolase family2 putative dipeptidase
69 d1wgoa_
72.2
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain70 c2c26A_
68.1
18
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: structural basis for the promiscuous specificity of the2 carbohydrate-binding modules from the beta-sandwich super3 family
71 c1uhvD_
not modelled
66.3
11
PDB header: hydrolaseChain: D: PDB Molecule: beta-xylosidase;PDBTitle: crystal structure of beta-d-xylosidase from2 thermoanaerobacterium saccharolyticum, a family 393 glycoside hydrolase
72 d1v6wa2
not modelled
65.6
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases73 d1tuxa_
not modelled
61.0
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases74 d1fh9a_
not modelled
57.1
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases75 d1auza_
not modelled
55.9
10
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Anti-sigma factor antagonist SpoIIaa76 d1xrsb2
not modelled
55.8
22
Fold: Dodecin subunit-likeSuperfamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domainFamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain77 c2xm5A_
not modelled
55.3
21
PDB header: transferaseChain: A: PDB Molecule: cloq;PDBTitle: structural and mechanistic analysis of the magnesium-2 independent aromatic prenyltransferase cloq from the3 clorobiocin biosynthetic pathway
78 d1qwga_
not modelled
52.1
17
Fold: TIM beta/alpha-barrelSuperfamily: (2r)-phospho-3-sulfolactate synthase ComAFamily: (2r)-phospho-3-sulfolactate synthase ComA79 d1bg4a_
not modelled
52.1
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases80 d1sfla_
not modelled
51.0
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase81 c3fdgA_
not modelled
49.9
17
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidase ac. metallo peptidase. merops family m19;PDBTitle: the crystal structure of the dipeptidase ac, metallo peptidase. merops2 family m19
82 c3jugA_
not modelled
45.3
12
PDB header: hydrolaseChain: A: PDB Molecule: beta-mannanase;PDBTitle: crystal structure of endo-beta-1,4-mannanase from the alkaliphilic2 bacillus sp. n16-5
83 c1l0qC_
not modelled
45.2
15
PDB header: protein bindingChain: C: PDB Molecule: surface layer protein;PDBTitle: tandem yvtn beta-propeller and pkd domains from an archaeal surface2 layer protein
84 d1w91a2
not modelled
43.5
9
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases85 c2wagA_
not modelled
42.8
11
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme, putative;PDBTitle: the structure of a family 25 glycosyl hydrolase from2 bacillus anthracis.
86 d1zdya1
not modelled
42.4
21
Fold: Antiparallel beta/alpha barrel (PT-barrel)Superfamily: Prenyltransferase-likeFamily: Prenyltransferase-like87 d1vc1a_
not modelled
41.8
13
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Anti-sigma factor antagonist SpoIIaa88 d1tv8a_
not modelled
41.1
12
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: MoCo biosynthesis proteins89 d1geha1
not modelled
41.0
20
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain90 c2kzwA_
not modelled
40.4
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of q8psa4 from methanosarcina mazei, northeast2 structural genomics consortium target mar143a
91 c2qw5B_
not modelled
40.3
12
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
92 d1itua_
not modelled
39.1
12
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: Renal dipeptidase93 d1ho8a_
not modelled
36.7
12
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Regulatory subunit H of the V-type ATPase94 c2o14A_
not modelled
35.1
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yxim;PDBTitle: x-ray crystal structure of protein yxim_bacsu from bacillus2 subtilis. northeast structural genomics consortium target3 sr595
95 c3emzA_
not modelled
34.8
13
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-xylanase;PDBTitle: crystal structure of xylanase xynb from paenibacillus2 barcinonensis complexed with a conduramine derivative
96 d1l0qa1
not modelled
34.4
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain97 c3itcA_
not modelled
33.9
13
PDB header: hydrolaseChain: A: PDB Molecule: renal dipeptidase;PDBTitle: crystal structure of sco3058 with bound citrate and glycerol
98 d1th8b_
not modelled
33.6
9
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Anti-sigma factor antagonist SpoIIaa99 d1ykwa1
not modelled
33.4
22
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain100 d1svda1
not modelled
32.6
16
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain101 c2xn1B_
not modelled
31.2
21
PDB header: hydrolaseChain: B: PDB Molecule: alpha-galactosidase;PDBTitle: structure of alpha-galactosidase from lactobacillus acidophilus ncfm2 with tris
102 d1v0la_
not modelled
30.9
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases103 c3jquA_
not modelled
30.8
12
PDB header: cell adhesionChain: A: PDB Molecule: collagenase;PDBTitle: crystal structure of clostridium histolyticum colg collagenase2 polycystic kidney disease domain at 1.4 angstrom resolution
104 c2qygC_
not modelled
30.7
18
PDB header: unknown functionChain: C: PDB Molecule: ribulose bisphosphate carboxylase-like protein 2;PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
105 d1rpxa_
not modelled
30.3
14
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase106 c3ndyA_
not modelled
29.8
15
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase d;PDBTitle: the structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
107 c2ylaA_
not modelled
29.7
14
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylhexosaminidase;PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
108 c2j75A_
not modelled
29.5
13
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucosidase a;PDBTitle: beta-glucosidase from thermotoga maritima in complex with2 noeuromycin
109 c2yr1B_
not modelled
29.5
16
PDB header: lyaseChain: B: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
110 c2zunB_
not modelled
29.1
15
PDB header: hydrolaseChain: B: PDB Molecule: 458aa long hypothetical endo-1,4-beta-glucanase;PDBTitle: functional analysis of hyperthermophilic endocellulase from2 the archaeon pyrococcus horikoshii
111 c3nsnA_
not modelled
28.4
16
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylglucosaminidase;PDBTitle: crystal structure of insect beta-n-acetyl-d-hexosaminidase ofhex12 complexed with tmg-chitotriomycin
112 d1q5ma_
not modelled
28.1
10
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)113 c3e77A_
not modelled
27.6
18
PDB header: transferaseChain: A: PDB Molecule: phosphoserine aminotransferase;PDBTitle: human phosphoserine aminotransferase in complex with plp
114 d1jaka1
not modelled
27.5
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain115 d1n82a_
not modelled
27.3
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases116 c2cksB_
not modelled
27.3
16
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase e-5;PDBTitle: x-ray crystal structure of the catalytic domain of2 thermobifida fusca endoglucanase cel5a (e5)
117 c3qm2A_
not modelled
27.1
23
PDB header: transferaseChain: A: PDB Molecule: phosphoserine aminotransferase;PDBTitle: 2.25 angstrom crystal structure of phosphoserine aminotransferase2 (serc) from salmonella enterica subsp. enterica serovar typhimurium
118 d1gqna_
not modelled
26.6
11
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase119 c2ou4C_
not modelled
26.3
8
PDB header: isomeraseChain: C: PDB Molecule: d-tagatose 3-epimerase;PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
120 c3sviA_
not modelled
25.5
22
PDB header: signaling proteinChain: A: PDB Molecule: type iii effector hopab2;PDBTitle: structure of the pto-binding domain of hoppmal generated by limited2 thermolysin digestion