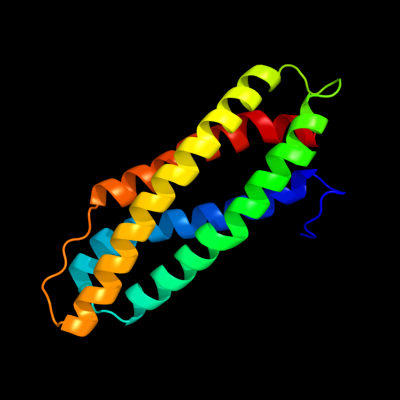
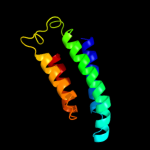
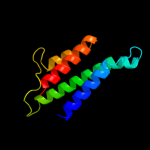
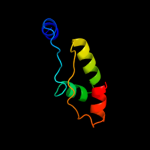
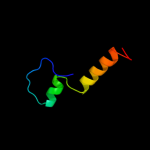
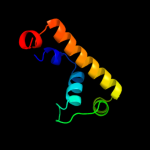
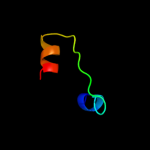
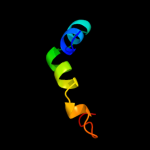
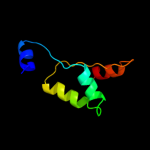
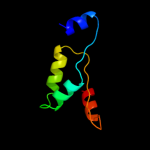
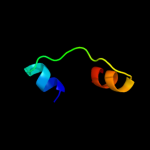
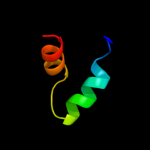
1 c2p0nA_
99.7
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein nmb1532;PDBTitle: nmb1532 protein from neisseria meningitidis, unknown function
2 c2k5eA_
99.6
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
3 c2k53A_
99.5
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: a3dk08 protein;PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
4 c3caxA_
98.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pf0695;PDBTitle: crystal structure of uncharacterized protein pf0695
5 d2fi0a1
97.3
14
Fold: SP0561-likeSuperfamily: SP0561-likeFamily: SP0561-like6 c2awyB_
93.9
14
PDB header: oxygen storage/transportChain: B: PDB Molecule: hemerythrin-like domain protein dcrh;PDBTitle: met-dcrh-hr
7 d2mhra_
72.7
17
Fold: Four-helical up-and-down bundleSuperfamily: Hemerythrin-likeFamily: Hemerythrin-like8 d2hmza_
71.7
16
Fold: Four-helical up-and-down bundleSuperfamily: Hemerythrin-likeFamily: Hemerythrin-like9 d2pt0a1
70.7
16
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Myo-inositol hexaphosphate phosphohydrolase (phytase) PhyA10 d1i4ya_
67.2
14
Fold: Four-helical up-and-down bundleSuperfamily: Hemerythrin-likeFamily: Hemerythrin-like11 d1hrba_
57.7
16
Fold: Four-helical up-and-down bundleSuperfamily: Hemerythrin-likeFamily: Hemerythrin-like12 d1qwga_
50.4
23
Fold: TIM beta/alpha-barrelSuperfamily: (2r)-phospho-3-sulfolactate synthase ComAFamily: (2r)-phospho-3-sulfolactate synthase ComA13 d1r9ja3
48.2
15
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Transketolase C-terminal domain-like14 d1ku7a_
47.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain15 c3o8nA_
39.7
27
PDB header: transferaseChain: A: PDB Molecule: 6-phosphofructokinase, muscle type;PDBTitle: structure of phosphofructokinase from rabbit skeletal muscle
16 d1d4ca3
39.0
6
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain17 c1u83A_
38.5
16
PDB header: lyaseChain: A: PDB Molecule: phosphosulfolactate synthase;PDBTitle: psl synthase from bacillus subtilis
18 d1u83a_
38.5
16
Fold: TIM beta/alpha-barrelSuperfamily: (2r)-phospho-3-sulfolactate synthase ComAFamily: (2r)-phospho-3-sulfolactate synthase ComA19 c2higA_
38.0
15
PDB header: transferaseChain: A: PDB Molecule: 6-phospho-1-fructokinase;PDBTitle: crystal structure of phosphofructokinase apoenzyme from trypanosoma2 brucei.
20 d1sm4a2
36.9
34
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases21 c2lfcA_
not modelled
35.3
15
PDB header: oxidoreductaseChain: A: PDB Molecule: fumarate reductase, flavoprotein subunit;PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
22 d2p5zx2
not modelled
35.0
15
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like23 d1y0pa3
not modelled
35.0
12
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain24 d4pfka_
not modelled
31.6
15
Fold: PhosphofructokinaseSuperfamily: PhosphofructokinaseFamily: Phosphofructokinase25 c3o8oB_
not modelled
30.5
23
PDB header: transferaseChain: B: PDB Molecule: 6-phosphofructokinase subunit beta;PDBTitle: structure of phosphofructokinase from saccharomyces cerevisiae
26 c2p9jH_
not modelled
29.8
13
PDB header: structural genomics, unknown functionChain: H: PDB Molecule: hypothetical protein aq2171;PDBTitle: crystal structure of aq2171 from aquifex aeolicus
27 d2djia2
not modelled
29.8
13
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module28 d2phcb1
not modelled
29.4
15
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like29 c3k2qA_
not modelled
29.3
27
PDB header: transferaseChain: A: PDB Molecule: pyrophosphate-dependent phosphofructokinase;PDBTitle: crystal structure of pyrophosphate-dependent2 phosphofructokinase from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr88
30 c3o8oC_
not modelled
29.3
27
PDB header: transferaseChain: C: PDB Molecule: 6-phosphofructokinase subunit alpha;PDBTitle: structure of phosphofructokinase from saccharomyces cerevisiae
31 d1pfka_
not modelled
29.1
20
Fold: PhosphofructokinaseSuperfamily: PhosphofructokinaseFamily: Phosphofructokinase32 c1zxxA_
not modelled
28.7
19
PDB header: transferaseChain: A: PDB Molecule: 6-phosphofructokinase;PDBTitle: the crystal structure of phosphofructokinase from lactobacillus2 delbrueckii
33 c2kebA_
not modelled
28.6
10
PDB header: dna binding proteinChain: A: PDB Molecule: dna polymerase subunit alpha b;PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
34 c2l1lB_
not modelled
27.8
16
PDB header: nuclear proteinChain: B: PDB Molecule: exportin-1;PDBTitle: nmr solution structure of the phi0 pki nes peptide in complex with2 crm1-rangtp
35 c1fncA_
not modelled
25.4
34
PDB header: oxidoreductase (nadp+(a),ferredoxin(a))Chain: A: PDB Molecule: ferredoxin-nadp+ reductase;PDBTitle: refined crystal structure of spinach ferredoxin reductase2 at 1.7 angstroms resolution: oxidized, reduced, and 2'-3 phospho-5'-amp bound states
36 c2p5zX_
not modelled
23.7
15
PDB header: structural genomics, unknown functionChain: X: PDB Molecule: type vi secretion system component;PDBTitle: the e. coli c3393 protein is a component of the type vi secretion2 system and exhibits structural similarity to t4 bacteriophage tail3 proteins gp27 and gp5
37 c3opyH_
not modelled
23.0
29
PDB header: transferaseChain: H: PDB Molecule: 6-phosphofructo-1-kinase beta-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
38 c3n1uA_
not modelled
22.6
5
PDB header: hydrolaseChain: A: PDB Molecule: hydrolase, had superfamily, subfamily iii a;PDBTitle: structure of putative had superfamily (subfamily iii a) hydrolase from2 legionella pneumophila
39 c3opyB_
not modelled
22.1
29
PDB header: transferaseChain: B: PDB Molecule: 6-phosphofructo-1-kinase beta-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
40 d1ffva1
not modelled
22.0
18
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like41 d1zxia1
not modelled
21.4
21
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like42 d1v97a1
not modelled
20.3
13
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like43 c3opyG_
not modelled
20.2
29
PDB header: transferaseChain: G: PDB Molecule: 6-phosphofructo-1-kinase alpha-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
44 d2f48a1
not modelled
20.2
19
Fold: PhosphofructokinaseSuperfamily: PhosphofructokinaseFamily: Phosphofructokinase45 d2al3a1
not modelled
19.8
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain46 c3g7pA_
not modelled
19.8
14
PDB header: unknown functionChain: A: PDB Molecule: nitrogen fixation protein;PDBTitle: crystal structure of a nifx-associated protein of unknown function2 (afe_1514) from acidithiobacillus ferrooxidans atcc at 2.00 a3 resolution
47 c3fiwB_
not modelled
19.7
20
PDB header: transcription regulatorChain: B: PDB Molecule: putative tetr-family transcriptional regulator;PDBTitle: structure of sco0253, a tetr-family transcriptional regulator from2 streptomyces coelicolor
48 d2piaa3
not modelled
18.6
4
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins49 d1ozha2
not modelled
18.1
7
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module50 d3e9va1
not modelled
17.8
15
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like51 d2z15a1
not modelled
17.6
18
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like52 c3hl4B_
not modelled
17.3
8
PDB header: transferaseChain: B: PDB Molecule: choline-phosphate cytidylyltransferase a;PDBTitle: crystal structure of a mammalian ctp:phosphocholine2 cytidylyltransferase with cdp-choline
53 c2rrdA_
not modelled
17.1
14
PDB header: dna binding proteinChain: A: PDB Molecule: hrdc domain from bloom syndrome protein;PDBTitle: structure of hrdc domain from human bloom syndrome protein, blm
54 c2pmzN_
not modelled
16.7
14
PDB header: translation, transferaseChain: N: PDB Molecule: dna-directed rna polymerase subunit n;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
55 d1qo8a3
not modelled
16.4
12
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain56 c3nj2B_
not modelled
16.4
9
PDB header: unknown functionChain: B: PDB Molecule: duf269-containing protein;PDBTitle: crystal structure of cce_0566 from the cyanobacterium cyanothece2 51142, a protein associated with nitrogen fixation from the duf2693 family
57 d1jt6a1
not modelled
16.1
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain58 d1d8ba_
not modelled
15.9
18
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases59 c2zp2B_
not modelled
15.6
23
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
60 d1n62a1
not modelled
15.5
18
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like61 d1wrua2
not modelled
14.9
26
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like62 d1f3ta2
not modelled
14.9
11
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain63 d1nd9a_
not modelled
14.7
20
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: N-terminal subdomain of bacterial translation initiation factor IF264 c3m8jA_
not modelled
14.5
7
PDB header: transcriptionChain: A: PDB Molecule: focb protein;PDBTitle: crystal structure of e.coli focb at 1.4 a resolution
65 c2xvcA_
not modelled
14.4
13
PDB header: cell cycleChain: A: PDB Molecule: escrt-iii;PDBTitle: molecular and structural basis of escrt-iii recruitment to2 membranes during archaeal cell division
66 d2p7vb1
not modelled
14.4
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain67 c3opyE_
not modelled
14.3
27
PDB header: transferaseChain: E: PDB Molecule: 6-phosphofructo-1-kinase alpha-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
68 c3b81A_
not modelled
14.0
13
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, acrr family;PDBTitle: crystal structure of predicted dna-binding transcriptional regulator2 of tetr/acrr family (np_350189.1) from clostridium acetobutylicum at3 2.10 a resolution
69 d1rm6c1
not modelled
14.0
13
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like70 c3he0A_
not modelled
13.6
13
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: the structure of a putative transcriptional regulator tetr family2 protein from vibrio parahaemolyticus.
71 c2wuiA_
not modelled
13.6
20
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of mexz, a key repressor responsible for2 antibiotic resistance in pseudomonas aeruginosa.
72 c2nvaH_
not modelled
13.6
6
PDB header: lyaseChain: H: PDB Molecule: arginine decarboxylase, a207r protein;PDBTitle: the x-ray crystal structure of the paramecium bursaria2 chlorella virus arginine decarboxylase bound to agmatine
73 c2qwwB_
not modelled
13.5
10
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
74 d1gaqb_
not modelled
13.4
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related75 d1pvda2
not modelled
13.3
18
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module76 d2i10a1
not modelled
13.1
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain77 c2b5oA_
not modelled
12.9
31
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin--nadp reductase;PDBTitle: ferredoxin-nadp reductase
78 c1pfuA_
not modelled
12.9
15
PDB header: ligaseChain: A: PDB Molecule: methionyl-trna synthetase;PDBTitle: methionyl-trna synthetase from escherichia coli complexed2 with methionine phosphinate
79 c3ccyA_
not modelled
12.8
13
PDB header: transcription regulatorChain: A: PDB Molecule: putative tetr-family transcriptional regulator;PDBTitle: crystal structure of a tetr-family transcriptional regulator from2 bordetella parapertussis 12822
80 d1gawa2
not modelled
12.7
28
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases81 d1losc_
not modelled
12.7
10
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase82 d2np5a1
not modelled
12.6
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain83 c3f6wE_
not modelled
12.5
13
PDB header: dna binding proteinChain: E: PDB Molecule: xre-family like protein;PDBTitle: xre-family like protein from pseudomonas syringae pv. tomato str.2 dc3000
84 c2qcoA_
not modelled
12.5
17
PDB header: transcriptionChain: A: PDB Molecule: cmer;PDBTitle: crystal structure of the transcriptional regulator cmer from2 campylobacter jejuni
85 d2cg4a1
not modelled
12.4
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain86 d3cdda2
not modelled
12.4
14
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like87 d1oe8a1
not modelled
12.3
22
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain88 d1h3na3
not modelled
12.3
13
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain89 d2fd5a1
not modelled
12.3
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain90 c3bcgA_
not modelled
12.2
20
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional regulator acrr;PDBTitle: conformational changes of the acrr regulator reveal a2 mechanism of induction
91 c2k29A_
not modelled
12.2
19
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
92 c2kwpA_
not modelled
12.2
21
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation protein nusa;PDBTitle: solution structure of the aminoterminal domain of e. coli nusa
93 d2g7la1
not modelled
12.1
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain94 d1pb6a1
not modelled
12.1
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain95 c3nbmA_
not modelled
12.1
28
PDB header: transferaseChain: A: PDB Molecule: pts system, lactose-specific iibc components;PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
96 c1wygA_
not modelled
12.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: xanthine dehydrogenase/oxidase;PDBTitle: crystal structure of a rat xanthine dehydrogenase triple mutant2 (c535a, c992r and c1324s)
97 c2dg7A_
not modelled
12.0
23
PDB header: gene regulationChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of the putative transcriptional regulator sco03372 from streptomyces coelicolor a3(2)
98 d4fxca_
not modelled
11.9
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related99 d2id3a1
not modelled
11.9
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain