1 c2wjyA_
100.0
19
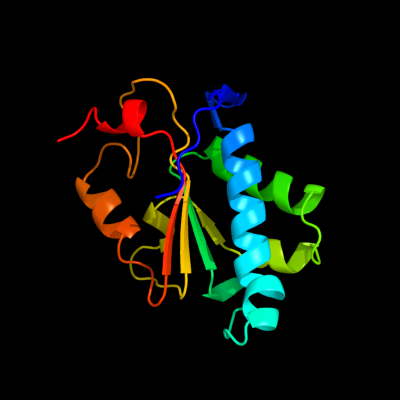
PDB header: hydrolaseChain: A: PDB Molecule: regulator of nonsense transcripts 1;PDBTitle: crystal structure of the complex between human nonsense2 mediated decay factors upf1 and upf2 orthorhombic form
2 c2xzlA_
100.0
23
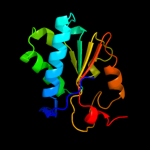
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent helicase nam7;PDBTitle: upf1-rna complex
3 c2gk7A_
100.0
19
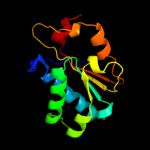
PDB header: hydrolaseChain: A: PDB Molecule: regulator of nonsense transcripts 1;PDBTitle: structural and functional insights into the human upf1 helicase core
4 c3hsiC_
99.9
11
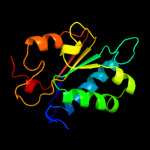
PDB header: transferaseChain: C: PDB Molecule: phosphatidylserine synthase;PDBTitle: crystal structure of phosphatidylserine synthase haemophilus2 influenzae rd kw20
5 d1byra_
99.8
16
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Nuclease6 d1v0wa2
99.8
16
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Phospholipase D7 c1v0sA_
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: phospholipase d;PDBTitle: uninhibited form of phospholipase d from streptomyces sp.2 strain pmf
8 d1xdpa3
99.8
22
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain9 d1v0wa1
99.8
15
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Phospholipase D10 c3e1sA_
99.6
20
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease v, subunit recd;PDBTitle: structure of an n-terminal truncation of deinococcus radiodurans recd2
11 c1w36G_
99.5
17
PDB header: recombinationChain: G: PDB Molecule: exodeoxyribonuclease v alpha chain;PDBTitle: recbcd:dna complex
12 c3lfuA_
99.4
16
PDB header: hydrolaseChain: A: PDB Molecule: dna helicase ii;PDBTitle: crystal structure of e. coli uvrd
13 c1pjrA_
99.3
19
PDB header: helicaseChain: A: PDB Molecule: pcra;PDBTitle: structure of dna helicase
14 c3dmnA_
99.2
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative dna helicase;PDBTitle: the crystal structure of the c-terminal domain of a possilbe dna2 helicase from lactobacillus plantarun wcfs1
15 c1xdoB_
99.1
16
PDB header: transferaseChain: B: PDB Molecule: polyphosphate kinase;PDBTitle: crystal structure of escherichia coli polyphosphate kinase
16 c2is6B_
99.0
21
PDB header: hydrolase/dnaChain: B: PDB Molecule: dna helicase ii;PDBTitle: crystal structure of uvrd-dna-adpmgf3 ternary complex
17 d1w36d2
99.0
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain18 c1uaaB_
98.9
16
PDB header: hydrolase/dnaChain: B: PDB Molecule: protein (atp-dependent dna helicase rep.);PDBTitle: e. coli rep helicase/dna complex
19 c2o8rA_
98.6
15
PDB header: transferaseChain: A: PDB Molecule: polyphosphate kinase;PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
20 d1xdpa4
98.5
20
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain21 d2o8ra3
not modelled
98.4
15
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain22 d2o8ra4
not modelled
98.2
15
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain23 c1qhhD_
not modelled
98.1
14
PDB header: hydrolaseChain: D: PDB Molecule: protein (pcra (subunit));PDBTitle: structure of dna helicase with adpnp
24 c2pjrB_
not modelled
97.9
18
PDB header: hydrolase/dnaChain: B: PDB Molecule: protein (helicase pcra);PDBTitle: helicase product complex
25 d1uaaa2
not modelled
97.9
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain26 d1w36b2
not modelled
97.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain27 d1pjra2
not modelled
97.7
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain28 c1w36E_
not modelled
97.1
19
PDB header: recombinationChain: E: PDB Molecule: exodeoxyribonuclease v beta chain;PDBTitle: recbcd:dna complex
29 c2c1lA_
not modelled
97.0
22
PDB header: hydrolaseChain: A: PDB Molecule: restriction endonuclease;PDBTitle: structure of the bfii restriction endonuclease
30 d1w36c2
not modelled
96.5
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain31 c1nopB_
not modelled
95.6
20
PDB header: hydrolase/dnaChain: B: PDB Molecule: tyrosyl-dna phosphodiesterase 1;PDBTitle: crystal structure of human tyrosyl-dna phosphodiesterase2 (tdp1) in complex with vanadate, dna and a human3 topoisomerase i-derived peptide
32 d1q32a2
not modelled
95.5
26
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Tyrosyl-DNA phosphodiesterase TDP133 d1jy1a2
not modelled
95.3
17
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Tyrosyl-DNA phosphodiesterase TDP134 c1w36F_
not modelled
94.8
13
PDB header: recombinationChain: F: PDB Molecule: exodeoxyribonuclease v gamma chain;PDBTitle: recbcd:dna complex
35 c1q32C_
not modelled
94.3
29
PDB header: replication,transcription,hydrolaseChain: C: PDB Molecule: tyrosyl-dna phosphodiesterase;PDBTitle: crystal structure analysis of the yeast tyrosyl-dna2 phosphodiesterase
36 c3sq3C_
not modelled
93.7
29
PDB header: hydrolaseChain: C: PDB Molecule: tyrosyl-dna phosphodiesterase 1;PDBTitle: crystal structure analysis of the yeast tyrosyl-dna phosphodiesterase2 h182a mutant
37 d2f5tx2
not modelled
91.3
13
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: TrmB middle domain-like38 c2pjrF_
not modelled
91.1
20
PDB header: hydrolase/dnaChain: F: PDB Molecule: protein (helicase pcra);PDBTitle: helicase product complex
39 c2f5tX_
not modelled
88.6
13
PDB header: transcriptionChain: X: PDB Molecule: archaeal transcriptional regulator trmb;PDBTitle: crystal structure of the sugar binding domain of the archaeal2 transcriptional regulator trmb
40 d1pzta_
not modelled
87.7
25
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: beta 1,4 galactosyltransferase (b4GalT1)41 c3lw6A_
not modelled
80.9
17
PDB header: transferaseChain: A: PDB Molecule: beta-4-galactosyltransferase 7;PDBTitle: crystal structure of drosophila beta1,4-galactosyltransferase-7
42 c1qhhA_
not modelled
66.1
13
PDB header: hydrolaseChain: A: PDB Molecule: protein (pcra (subunit));PDBTitle: structure of dna helicase with adpnp
43 d1jy1a1
not modelled
57.9
17
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Tyrosyl-DNA phosphodiesterase TDP144 c1t3gB_
not modelled
33.0
9
PDB header: membrane proteinChain: B: PDB Molecule: x-linked interleukin-1 receptor accessoryPDBTitle: crystal structure of the toll/interleukin-1 receptor (tir)2 domain of human il-1rapl
45 c1qhhB_
not modelled
29.4
21
PDB header: hydrolaseChain: B: PDB Molecule: protein (pcra (subunit));PDBTitle: structure of dna helicase with adpnp
46 d1gz0a2
not modelled
23.9
15
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: RNA 2'-O ribose methyltransferase substrate binding domain47 d1s2da_
not modelled
23.6
21
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase48 d1f8ya_
not modelled
22.1
13
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase49 c3oziB_
not modelled
21.9
13
PDB header: plant proteinChain: B: PDB Molecule: l6tr;PDBTitle: crystal structure of the tir domain from the flax disease resistance2 protein l6
50 d1qzqa1
not modelled
20.9
19
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Tyrosyl-DNA phosphodiesterase TDP151 c3fniA_
not modelled
17.8
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
52 c3dy0B_
not modelled
17.4
33
PDB header: blood clotting, hydrolase inhibitorChain: B: PDB Molecule: c-terminus plasma serine protease inhibitor;PDBTitle: crystal structure of cleaved pci bound to heparin
53 c3p1wA_
not modelled
16.6
20
PDB header: protein transportChain: A: PDB Molecule: rabgdi protein;PDBTitle: crystal structure of rab gdi from plasmodium falciparum, pfl2060c
54 d1q32a1
not modelled
16.5
24
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Tyrosyl-DNA phosphodiesterase TDP155 c1lq8H_
not modelled
16.3
33
PDB header: blood clottingChain: H: PDB Molecule: plasma serine protease inhibitor;PDBTitle: crystal structure of cleaved protein c inhibitor
56 c3cgiD_
not modelled
16.2
21
PDB header: unknown functionChain: D: PDB Molecule: propanediol utilization protein pduu;PDBTitle: crystal structure of the pduu shell protein from the pdu2 microcompartment
57 d1gz0f2
not modelled
15.8
15
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: RNA 2'-O ribose methyltransferase substrate binding domain58 c3mpbA_
not modelled
15.6
24
PDB header: isomeraseChain: A: PDB Molecule: sugar isomerase;PDBTitle: z5688 from e. coli o157:h7 bound to fructose
59 d1o98a1
not modelled
14.5
18
Fold: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domainSuperfamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domainFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain60 c3pstA_
not modelled
12.6
20
PDB header: nuclear proteinChain: A: PDB Molecule: protein doa1;PDBTitle: crystal structure of pul and pfu(mutate) domain
61 d1r89a1
not modelled
12.5
24
Fold: PAP/OAS1 substrate-binding domainSuperfamily: PAP/OAS1 substrate-binding domainFamily: Archaeal tRNA CCA-adding enzyme substrate-binding domain62 c3ia0c_
not modelled
12.4
18
PDB header: structural proteinChain: C: PDB Molecule: ethanolamine utilization protein euts;PDBTitle: ethanolamine utilization microcompartment shell subunit,2 euts-g39v mutant
63 c3fd0B_
not modelled
12.1
12
PDB header: lyaseChain: B: PDB Molecule: putative cystathionine beta-lyase involved in aluminumPDBTitle: crystal structure of putative cystathionine beta-lyase involved in2 aluminum resistance (np_470671.1) from listeria innocua at 2.12 a3 resolution
64 c3l3fX_
not modelled
12.1
20
PDB header: protein bindingChain: X: PDB Molecule: protein doa1;PDBTitle: crystal structure of a pfu-pul domain pair of saccharomyces cerevisiae2 doa1/ufd3
65 c2l82A_
not modelled
12.1
14
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or32;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or32
66 c2a5hC_
not modelled
12.0
8
PDB header: isomeraseChain: C: PDB Molecule: l-lysine 2,3-aminomutase;PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
67 d1xo1a2
not modelled
12.0
15
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain68 c3ktbD_
not modelled
11.5
19
PDB header: transcription regulatorChain: D: PDB Molecule: arsenical resistance operon trans-acting repressor;PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
69 c2y0oA_
not modelled
10.9
28
PDB header: isomeraseChain: A: PDB Molecule: probable d-lyxose ketol-isomerase;PDBTitle: the structure of a d-lyxose isomerase from the sigmab2 regulon of bacillus subtilis
70 c2z86D_
not modelled
10.8
15
PDB header: transferaseChain: D: PDB Molecule: chondroitin synthase;PDBTitle: crystal structure of chondroitin polymerase from2 escherichia coli strain k4 (k4cp) complexed with udp-glcua3 and udp
71 d1utaa_
not modelled
9.5
21
Fold: Ferredoxin-likeSuperfamily: Sporulation related repeatFamily: Sporulation related repeat72 c3ckvA_
not modelled
9.5
16
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a mycobacterial protein
73 d1x94a_
not modelled
8.9
17
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain74 d1vmea1
not modelled
8.8
17
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related75 d1j5pa4
not modelled
8.7
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain76 c3kgkA_
not modelled
8.5
15
PDB header: chaperoneChain: A: PDB Molecule: arsenical resistance operon trans-acting repressor arsd;PDBTitle: crystal structure of arsd
77 c2r60A_
not modelled
8.4
9
PDB header: transferaseChain: A: PDB Molecule: glycosyl transferase, group 1;PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
78 c3iabB_
not modelled
8.3
25
PDB header: hydrolase/rnaChain: B: PDB Molecule: ribonucleases p/mrp protein subunit pop7;PDBTitle: crystal structure of rnase p /rnase mrp proteins pop6, pop72 in a complex with the p3 domain of rnase mrp rna
79 c2bcwC_
not modelled
8.2
13
PDB header: ribosomeChain: C: PDB Molecule: elongation factor g;PDBTitle: coordinates of the n-terminal domain of ribosomal protein2 l11,c-terminal domain of ribosomal protein l7/l12 and a3 portion of the g' domain of elongation factor g, as fitted4 into cryo-em map of an escherichia coli 70s*ef-5 g*gdp*fusidic acid complex
80 c2x0dA_
not modelled
7.9
7
PDB header: transferaseChain: A: PDB Molecule: wsaf;PDBTitle: apo structure of wsaf
81 d1o0sa2
not modelled
7.6
12
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Malic enzyme N-domain82 d1fyva_
not modelled
7.5
11
Fold: Flavodoxin-likeSuperfamily: Toll/Interleukin receptor TIR domainFamily: Toll/Interleukin receptor TIR domain83 c2zrrA_
not modelled
7.5
30
PDB header: antimicrobial proteinChain: A: PDB Molecule: mundticin ks immunity protein;PDBTitle: crystal structure of an immunity protein that contributes2 to the self-protection of bacteriocin-producing3 enterococcus mundtii 15-1a
84 d1w36d1
not modelled
7.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain85 c3h16A_
not modelled
6.1
16
PDB header: signaling proteinChain: A: PDB Molecule: tir protein;PDBTitle: crystal structure of a bacteria tir domain, pdtir from2 paracoccus denitrificans
86 c2dc1A_
not modelled
6.1
13
PDB header: oxidoreductaseChain: A: PDB Molecule: l-aspartate dehydrogenase;PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
87 c3cseA_
not modelled
6.0
32
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrofolate reductase;PDBTitle: candida glabrata dihydrofolate reductase complexed with2 nadph and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-3 ynyl)-6-ethylpyrimidine (ucp120b)
88 c3nklA_
not modelled
5.9
15
PDB header: oxidoreductase/lyaseChain: A: PDB Molecule: udp-d-quinovosamine 4-dehydrogenase;PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
89 c3cvjB_
not modelled
5.7
17
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
90 c2ihnA_
not modelled
5.5
15
PDB header: hydrolase/dnaChain: A: PDB Molecule: ribonuclease h;PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
91 c3ht4B_
not modelled
5.5
14
PDB header: lyaseChain: B: PDB Molecule: aluminum resistance protein;PDBTitle: crystal structure of the q81a77_baccr protein from bacillus2 cereus. northeast structural genomics consortium target3 bcr213
92 c3nkuA_
not modelled
5.5
20
PDB header: protein transportChain: A: PDB Molecule: drra;PDBTitle: crystal structure of the n-terminal domain of drra/sidm from2 legionella pneumophila
93 c2k19A_
not modelled
5.4
30
PDB header: antimicrobial proteinChain: A: PDB Molecule: putative piscicolin 126 immunity protein;PDBTitle: nmr solution structure of pisi
94 c2bpbB_
not modelled
5.3
20
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
95 c1o98A_
not modelled
5.2
18
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
96 c2x3yA_
not modelled
5.2
20
PDB header: isomeraseChain: A: PDB Molecule: phosphoheptose isomerase;PDBTitle: crystal structure of gmha from burkholderia pseudomallei