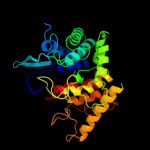
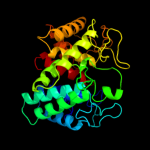
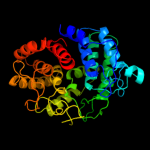
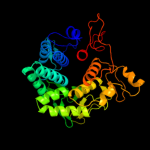
1 c3qxqD_
100.0
100
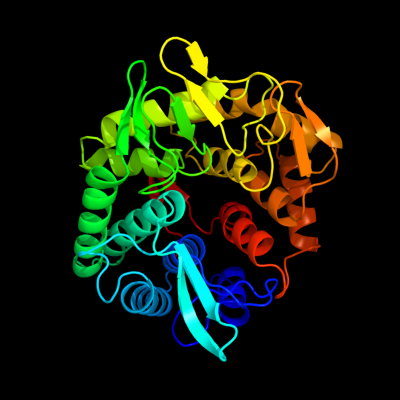
PDB header: hydrolaseChain: D: PDB Molecule: endoglucanase;PDBTitle: structure of the bacterial cellulose synthase subunit z in complex2 with cellopentaose
2 d1wzza1
100.0
26
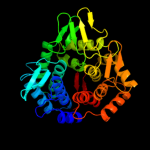
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain3 d1wu4a1
100.0
22
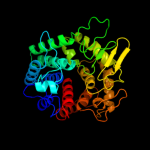
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain4 d1h12a_
100.0
22
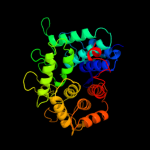
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain5 d1kwfa_
100.0
22
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain6 d1v5da_
100.0
18
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain7 c3renB_
100.0
14
PDB header: hydrolaseChain: B: PDB Molecule: glycosyl hydrolase, family 8;PDBTitle: cpf_2247, a novel alpha-amylase from clostridium perfringens
8 c2zzrA_
94.2
14
PDB header: hydrolaseChain: A: PDB Molecule: unsaturated glucuronyl hydrolase;PDBTitle: crystal structure of unsaturated glucuronyl hydrolase from2 streptcoccus agalactiae
9 d1fp3a_
93.9
12
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: N-acylglucosamine (NAG) epimerase10 d2afaa1
93.3
8
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: N-acylglucosamine (NAG) epimerase11 c3pmmA_
90.2
11
PDB header: hydrolaseChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: the crystal structure of a possible member of gh105 family from2 klebsiella pneumoniae subsp. pneumoniae mgh 78578
12 c3k11A_
88.9
7
PDB header: hydrolaseChain: A: PDB Molecule: putative glycosyl hydrolase;PDBTitle: crystal structure of putative glycosyl hydrolase (np_813087.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.80 a resolution
13 d1nxca_
83.2
15
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain14 c3k7xA_
74.6
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin0763 protein;PDBTitle: crystal structure of the lin0763 protein from listeria2 innocua. northeast structural genomics consortium target3 lkr23.
15 c3eu8D_
69.7
14
PDB header: hydrolaseChain: D: PDB Molecule: putative glucoamylase;PDBTitle: crystal structure of putative glucoamylase (yp_210071.1) from2 bacteroides fragilis nctc 9343 at 2.12 a resolution
16 c3qspB_
68.0
15
PDB header: hydrolaseChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: analysis of a new family of widely distributed metal-independent alpha2 mannosidases provides unique insight into the processing of n-linked3 glycans, streptococcus pneumoniae sp_2144 non-productive substrate4 complex with alpha-1,6-mannobiose
17 d1g9ga_
55.3
30
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain18 c1x9dA_
47.2
14
PDB header: hydrolaseChain: A: PDB Molecule: endoplasmic reticulum mannosyl-oligosaccharide 1,PDBTitle: crystal structure of human class i alpha-1,2-mannosidase in2 complex with thio-disaccharide substrate analogue
19 d1x9da1
47.2
14
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain20 c2gz6B_
42.9
7
PDB header: isomeraseChain: B: PDB Molecule: n-acetyl-d-glucosamine 2-epimerase;PDBTitle: crystal structure of anabaena sp. ch1 n-acetyl-d-glucosamine 2-2 epimerase at 2.0 a
21 c3e6uA_
not modelled
37.6
14
PDB header: signaling proteinChain: A: PDB Molecule: lanc-like protein 1;PDBTitle: crystal structure of human lancl1
22 d1hcua_
not modelled
34.1
12
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain23 d2p0va1
not modelled
31.2
14
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: CPF0428-like24 c2p0vA_
not modelled
31.2
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein bt3781;PDBTitle: crystal structure of bt3781 protein from bacteroides2 thetaiotaomicron, northeast structural genomics target3 btr58
25 c3okfA_
not modelled
21.6
24
PDB header: lyaseChain: A: PDB Molecule: 3-dehydroquinate synthase;PDBTitle: 2.5 angstrom resolution crystal structure of 3-dehydroquinate synthase2 (arob) from vibrio cholerae
26 d2g0da1
not modelled
18.0
14
Fold: alpha/alpha toroidSuperfamily: LanC-likeFamily: LanC-like27 c3rbbA_
not modelled
17.4
30
PDB header: viral protein, protein bindingChain: A: PDB Molecule: protein nef;PDBTitle: hiv-1 nef protein in complex with engineered hck sh3 domain
28 d2nvpa1
not modelled
16.9
14
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: CPF0428-like29 d1l1ya_
not modelled
15.8
16
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain30 c1l2aD_
not modelled
15.8
16
PDB header: hydrolaseChain: D: PDB Molecule: cellobiohydrolase;PDBTitle: the crystal structure and catalytic mechanism of2 cellobiohydrolase cels, the major enzymatic component of3 the clostridium thermocellum cellulosome
31 d2nefa_
not modelled
15.8
29
Fold: Regulatory factor NefSuperfamily: Regulatory factor NefFamily: Regulatory factor Nef32 c3jtgA_
not modelled
15.7
23
PDB header: transcriptionChain: A: PDB Molecule: ets-related transcription factor elf-3;PDBTitle: crystal structure of mouse elf3 c-terminal dna-binding domain in2 complex with type ii tgf-beta receptor promoter dna
33 c3gt5A_
not modelled
15.1
10
PDB header: isomeraseChain: A: PDB Molecule: n-acetylglucosamine 2-epimerase;PDBTitle: crystal structure of an n-acetylglucosamine 2-epimerase family protein2 from xylella fastidiosa
34 d1nc5a_
not modelled
13.7
14
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Hypothetical protein YteR35 c1p0oA_
not modelled
13.0
30
PDB header: ribosomeChain: A: PDB Molecule: 19-mer peptide from 50s ribosomal protein l1;PDBTitle: hp (2-20) substitution of trp for gln and asp at position2 17 and 19 modification in sds-d25 micelles
36 c3l55B_
not modelled
12.7
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: b-1,4-endoglucanase/cellulase;PDBTitle: crystal structure of a putative beta-1,4-endoglucanase /2 cellulase from prevotella bryantii
37 c3ik5A_
not modelled
12.6
25
PDB header: viral protein/signaling proteinChain: A: PDB Molecule: protein nef;PDBTitle: sivmac239 nef in complex with tcr zeta itam 1 polypeptide2 (a63-r80)
38 c2d3wB_
not modelled
12.1
29
PDB header: biosynthetic proteinChain: B: PDB Molecule: probable atp-dependent transporter sufc;PDBTitle: crystal structure of escherichia coli sufc, an atpase2 compenent of the suf iron-sulfur cluster assembly machinery
39 c2dcoA_
not modelled
10.8
63
PDB header: membrane proteinChain: A: PDB Molecule: s1p4 first extracellular loop peptidomimetic;PDBTitle: s1p4 first extracellular loop peptidomimetic
40 c2xi1A_
not modelled
10.0
30
PDB header: viral proteinChain: A: PDB Molecule: nef;PDBTitle: crystal structure of the hiv-1 nef sequenced from a patient's sample
41 d1jb7a1
not modelled
9.8
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB42 c3nfvA_
not modelled
9.0
13
PDB header: lyaseChain: A: PDB Molecule: alginate lyase;PDBTitle: crystal structure of an alginate lyase (bacova_01668) from bacteroides2 ovatus at 1.95 a resolution
43 d1efnb_
not modelled
8.9
30
Fold: Regulatory factor NefSuperfamily: Regulatory factor NefFamily: Regulatory factor Nef44 d1puee_
not modelled
8.4
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ets domain45 d1lm7a_
not modelled
8.3
20
Fold: beta-hairpin-alpha-hairpin repeatSuperfamily: Plakin repeatFamily: Plakin repeat46 c2oylB_
not modelled
7.9
15
PDB header: hydrolaseChain: B: PDB Molecule: endoglycoceramidase ii;PDBTitle: endo-glycoceramidase ii from rhodococcus sp.: cellobiose-like2 imidazole complex
47 d1rmva_
not modelled
6.8
24
Fold: Four-helical up-and-down bundleSuperfamily: TMV-like viral coat proteinsFamily: TMV-like viral coat proteins48 d1g99a2
not modelled
6.6
31
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like49 d1pgl21
not modelled
6.6
19
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP50 d1pgw21
not modelled
6.5
19
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP51 d1ny721
not modelled
6.4
13
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP52 d1xjva1
not modelled
6.3
12
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB53 d1ei7a_
not modelled
6.2
21
Fold: Four-helical up-and-down bundleSuperfamily: TMV-like viral coat proteinsFamily: TMV-like viral coat proteins54 c1lf6A_
not modelled
6.1
20
PDB header: hydrolaseChain: A: PDB Molecule: glucoamylase;PDBTitle: crystal structure of bacterial glucoamylase
55 c3pdmP_
not modelled
6.1
24
PDB header: virusChain: P: PDB Molecule: coat protein;PDBTitle: hibiscus latent singapore virus
56 d2d5ja1
not modelled
5.9
10
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Glycosyl Hydrolase Family 8857 d1vtmp_
not modelled
5.9
17
Fold: Four-helical up-and-down bundleSuperfamily: TMV-like viral coat proteinsFamily: TMV-like viral coat proteins58 c3sm3A_
not modelled
5.9
12
PDB header: transferaseChain: A: PDB Molecule: sam-dependent methyltransferases;PDBTitle: crystal structure of sam-dependent methyltransferases q8puk2_metma2 from methanosarcina mazei. northeast structural genomics consortium3 target mar262.
59 c3mtiA_
not modelled
5.7
19
PDB header: transferaseChain: A: PDB Molecule: rrna methylase;PDBTitle: the crystal structure of a rrna methylase from streptococcus2 thermophilus to 1.95a
60 c2bfuL_
not modelled
5.6
10
PDB header: virusChain: L: PDB Molecule: cowpea mosaic virus, large (l) subunit;PDBTitle: x-ray structure of cpmv top component
61 c1bmv2_
not modelled
5.4
14
PDB header: virus/rnaChain: 2: PDB Molecule: protein (icosahedral virus - b and c domain);PDBTitle: protein-rna interactions in an icosahedral virus at 3.02 angstroms resolution
62 c1ta9A_
not modelled
5.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol dehydrogenase;PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
63 c2ov2O_
not modelled
5.1
29
PDB header: protein binding/transferaseChain: O: PDB Molecule: serine/threonine-protein kinase pak 4;PDBTitle: the crystal structure of the human rac3 in complex with the crib2 domain of human p21-activated kinase 4 (pak4)
64 c2yfvC_
not modelled
5.0
35
PDB header: cell cycleChain: C: PDB Molecule: scm3;PDBTitle: the heterotrimeric complex of kluyveromyces lactis scm3, cse4 and h4