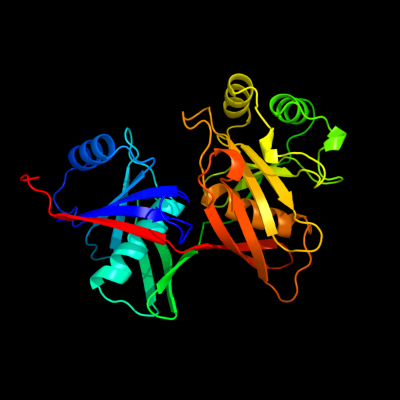
| 1 | c1qy9B_
|
|
 |
100.0 |
98 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein ydde;
PDBTitle: crystal structure of e. coli se-met protein ydde
|
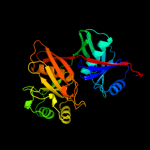
| 2 | c3ednB_
|
|
 |
100.0 |
32 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:phenazine biosynthesis protein, phzf family;
PDBTitle: crystal structure of the bacillus anthracis phenazine2 biosynthesis protein, phzf family
|
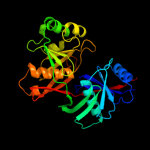
| 3 | c1ym5A_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical 32.6 kda protein in dap2-slt2
PDBTitle: crystal structure of yhi9, the yeast member of the2 phenazine biosynthesis phzf enzyme superfamily.
|
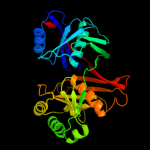
| 4 | c1u1wA_
|
|
 |
100.0 |
30 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:phenazine biosynthesis protein phzf;
PDBTitle: structure and function of phenazine-biosynthesis protein phzf from2 pseudomonas fluorescens 2-79
|
| 5 | c1u0kA_
|
|
 |
100.0 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:gene product pa4716;
PDBTitle: the structure of a predicted epimerase pa4716 from pseudomonas2 aeruginosa
|
| 6 | d1s7ja_
|
|
 |
100.0 |
31 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 7 | c3ekmE_
|
|
 |
100.0 |
16 |
PDB header:isomerase
Chain: E: PDB Molecule:diaminopimelate epimerase, chloroplastic;
PDBTitle: crystal structure of diaminopimelate epimerase form2 arabidopsis thaliana in complex with irreversible inhibitor3 dl-azidap
|
| 8 | c2otnB_
|
|
 |
100.0 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of the catalytically active form of diaminopimelate2 epimerase from bacillus anthracis
|
| 9 | c2gkjA_
|
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of diaminopimelate epimerase in complex2 with an irreversible inhibitor dl-azidap
|
| 10 | c2azpA_
|
|
 |
100.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pa1268;
PDBTitle: crystal structure of pa1268 solved by sulfur sad
|
| 11 | c1w62B_
|
|
 |
100.0 |
13 |
PDB header:racemase
Chain: B: PDB Molecule:b-cell mitogen;
PDBTitle: proline racemase in complex with one molecule of pyrrole-2-2 carboxylic acid (hemi form)
|
| 12 | d1qy9a1
|
|
 |
100.0 |
99 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 13 | c3fveA_
|
|
 |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of diaminopimelate epimerase mycobacterium2 tuberculosis dapf
|
| 14 | d1u0ka1
|
|
 |
100.0 |
26 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 15 | d1xuba1
|
|
 |
100.0 |
33 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 16 | d1qy9a2
|
|
 |
100.0 |
98 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 17 | d1tm0a_
|
|
 |
100.0 |
14 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Proline racemase |
| 18 | d1xuba2
|
|
 |
100.0 |
30 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 19 | d1u0ka2
|
|
 |
99.9 |
30 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 20 | d2gkea1
|
|
 |
99.9 |
20 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Diaminopimelate epimerase |
| 21 | d2gkea2 |
|
not modelled |
99.8 |
13 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Diaminopimelate epimerase |
| 22 | c3g7kD_ |
|
not modelled |
98.2 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:3-methylitaconate isomerase;
PDBTitle: crystal structure of methylitaconate-delta-isomerase
|
| 23 | c2pw0A_ |
|
not modelled |
98.1 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:prpf methylaconitate isomerase;
PDBTitle: crystal structure of trans-aconitate bound to methylaconitate2 isomerase prpf from shewanella oneidensis
|
| 24 | d2h9fa2 |
|
not modelled |
96.0 |
23 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PA0793-like |
| 25 | c3ldgA_ |
|
not modelled |
40.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein smu.472;
PDBTitle: crystal structure of smu.472, a putative methyltransferase complexed2 with sah
|
| 26 | c3k0bA_ |
|
not modelled |
38.8 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted n6-adenine-specific dna methylase;
PDBTitle: crystal structure of a predicted n6-adenine-specific dna methylase2 from listeria monocytogenes str. 4b f2365
|
| 27 | d1g60a_ |
|
not modelled |
36.5 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Type II DNA methylase |
| 28 | d1booa_ |
|
not modelled |
33.8 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Type II DNA methylase |
| 29 | c2zifB_ |
|
not modelled |
32.7 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:putative modification methylase;
PDBTitle: crystal structure of ttha0409, putative dna modification2 methylase from thermus thermophilus hb8- complexed with s-3 adenosyl-l-methionine
|
| 30 | d1euza2 |
|
not modelled |
31.7 |
26 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 31 | d1bgva2 |
|
not modelled |
31.5 |
19 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 32 | d1v9la2 |
|
not modelled |
31.0 |
19 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 33 | c1nw6A_ |
|
not modelled |
29.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:modification methylase rsri;
PDBTitle: structure of the beta class n6-adenine dna methyltransferase rsri2 bound to sinefungin
|
| 34 | d1o9ga_ |
|
not modelled |
28.7 |
27 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:rRNA methyltransferase AviRa |
| 35 | d1gtma2 |
|
not modelled |
26.6 |
23 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 36 | d1eg2a_ |
|
not modelled |
24.2 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Type II DNA methylase |
| 37 | c3lduA_ |
|
not modelled |
23.6 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:putative methylase;
PDBTitle: the crystal structure of a possible methylase from2 clostridium difficile 630.
|
| 38 | d1bvua2 |
|
not modelled |
23.4 |
23 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 39 | d2ar0a1 |
|
not modelled |
22.3 |
36 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:N-6 DNA Methylase-like |
| 40 | d2fug21 |
|
not modelled |
21.7 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:NQO2-like |
| 41 | d1b26a2 |
|
not modelled |
21.7 |
26 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 42 | c2waqG_ |
|
not modelled |
20.6 |
27 |
PDB header:transcription
Chain: G: PDB Molecule:dna-directed rna polymerase rpo8 subunit;
PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
|
| 43 | c3khkA_ |
|
not modelled |
20.2 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:type i restriction-modification system
PDBTitle: crystal structure of type-i restriction-modification system2 methylation subunit (mm_0429) from methanosarchina mazei.
|
| 44 | d1hwxa2 |
|
not modelled |
19.3 |
29 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 45 | d2okca1 |
|
not modelled |
17.8 |
29 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:N-6 DNA Methylase-like |
| 46 | c2l3bA_ |
|
not modelled |
17.4 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein found in conjugate transposon;
PDBTitle: solution nmr structure of the bt_0084 lipoprotein from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr376
|
| 47 | d2f8la1 |
|
not modelled |
17.2 |
36 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:N-6 DNA Methylase-like |
| 48 | c2l7qA_ |
|
not modelled |
16.3 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein found in conjugate transposon;
PDBTitle: solution nmr structure of conjugate transposon protein bvu_1572(27-2 141) from bacteroides vulgatus, northeast structural genomics3 consortium target bvr155
|
| 49 | c3p9nA_ |
|
not modelled |
16.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:possible methyltransferase (methylase);
PDBTitle: rv2966c of m. tuberculosis is a rsmd-like methyltransferase
|
| 50 | c1g38A_ |
|
not modelled |
15.4 |
7 |
PDB header:transferase/dna
Chain: A: PDB Molecule:modification methylase taqi;
PDBTitle: adenine-specific methyltransferase m. taq i/dna complex
|
| 51 | c3sb1B_ |
|
not modelled |
15.4 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hydrogenase expression protein;
PDBTitle: hydrogenase expression protein huph from thiobacillus denitrificans2 atcc 25259
|
| 52 | c1aqjB_ |
|
not modelled |
15.4 |
7 |
PDB header:methyltransferase
Chain: B: PDB Molecule:adenine-n6-dna-methyltransferase taqi;
PDBTitle: structure of adenine-n6-dna-methyltransferase taqi
|
| 53 | d1uhsa_ |
|
not modelled |
15.2 |
35 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 54 | d2ih2a1 |
|
not modelled |
14.0 |
7 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:DNA methylase TaqI, N-terminal domain |
| 55 | d1l1fa2 |
|
not modelled |
14.0 |
29 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 56 | c3lkdB_ |
|
not modelled |
13.5 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:type i restriction-modification system
PDBTitle: crystal structure of the type i restriction-modification2 system methyltransferase subunit from streptococcus3 thermophilus, northeast structural genomics consortium4 target sur80
|
| 57 | d1s7ea1 |
|
not modelled |
13.2 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 58 | d1j6ua1 |
|
not modelled |
13.1 |
12 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 59 | d1ko7a1 |
|
not modelled |
13.1 |
13 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:HPr kinase/phoshatase HprK N-terminal domain |
| 60 | d2p81a1 |
|
not modelled |
12.2 |
41 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 61 | c3itcA_ |
|
not modelled |
12.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:renal dipeptidase;
PDBTitle: crystal structure of sco3058 with bound citrate and glycerol
|
| 62 | c3k8zD_ |
|
not modelled |
11.8 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nad-specific glutamate dehydrogenase;
PDBTitle: crystal structure of gudb1 a decryptified secondary glutamate2 dehydrogenase from b. subtilis
|
| 63 | d1pjza_ |
|
not modelled |
11.7 |
24 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Thiopurine S-methyltransferase |
| 64 | c3c6vB_ |
|
not modelled |
11.2 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:probable tautomerase/dehalogenase au4130;
PDBTitle: crystal structure of au4130/apc7354, a probable enzyme from the2 thermophilic fungus aspergillus fumigatus
|
| 65 | c1v9lA_ |
|
not modelled |
11.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase;
PDBTitle: l-glutamate dehydrogenase from pyrobaculum islandicum2 complexed with nad
|
| 66 | d1e3oc1 |
|
not modelled |
11.0 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 67 | d1knxa1 |
|
not modelled |
10.9 |
3 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:HPr kinase/phoshatase HprK N-terminal domain |
| 68 | d1wy7a1 |
|
not modelled |
10.8 |
33 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Ta1320-like |
| 69 | c2i5gB_ |
|
not modelled |
10.6 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:amidohydrolase;
PDBTitle: crystal strcuture of amidohydrolase from pseudomonas2 aeruginosa
|
| 70 | c1bvuF_ |
|
not modelled |
10.4 |
23 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:protein (glutamate dehydrogenase);
PDBTitle: glutamate dehydrogenase from thermococcus litoralis
|
| 71 | c2vi6F_ |
|
not modelled |
10.3 |
41 |
PDB header:transcription
Chain: F: PDB Molecule:homeobox protein nanog;
PDBTitle: crystal structure of the nanog homeodomain
|
| 72 | c2da4A_ |
|
not modelled |
10.3 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein dkfzp686k21156;
PDBTitle: solution structure of the homeobox domain of the2 hypothetical protein, dkfzp686k21156
|
| 73 | c3lu2B_ |
|
not modelled |
10.2 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2462 protein;
PDBTitle: structure of lmo2462, a listeria monocytogenes amidohydrolase family2 putative dipeptidase
|
| 74 | d1o4xa1 |
|
not modelled |
10.1 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 75 | c3s1sA_ |
|
not modelled |
10.0 |
29 |
PDB header:hydrolase, transferase
Chain: A: PDB Molecule:restriction endonuclease bpusi;
PDBTitle: characterization and crystal structure of the type iig restriction2 endonuclease bpusi
|
| 76 | d1p7ia_ |
|
not modelled |
9.8 |
41 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 77 | c2da2A_ |
|
not modelled |
9.7 |
5 |
PDB header:transcription
Chain: A: PDB Molecule:alpha-fetoprotein enhancer binding protein;
PDBTitle: solution structure of the second homeobox domain of at-2 binding transcription factor 1 (atbf1)
|
| 78 | c2tmgD_ |
|
not modelled |
9.5 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:protein (glutamate dehydrogenase);
PDBTitle: thermotoga maritima glutamate dehydrogenase mutant s128r,2 t158e, n117r, s160e
|
| 79 | d1ne2a_ |
|
not modelled |
9.4 |
36 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Ta1320-like |
| 80 | d2hi3a1 |
|
not modelled |
9.3 |
35 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 81 | d1ws6a1 |
|
not modelled |
9.3 |
29 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
| 82 | c2da1A_ |
|
not modelled |
9.2 |
29 |
PDB header:transcription
Chain: A: PDB Molecule:alpha-fetoprotein enhancer binding protein;
PDBTitle: solution structure of the first homeobox domain of at-2 binding transcription factor 1 (atbf1)
|
| 83 | d2ifta1 |
|
not modelled |
9.0 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
| 84 | c2yfqA_ |
|
not modelled |
9.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad-specific glutamate dehydrogenase;
PDBTitle: crystal structure of glutamate dehydrogenase from2 peptoniphilus asaccharolyticus
|
| 85 | c2l9rA_ |
|
not modelled |
8.9 |
29 |
PDB header:transcription
Chain: A: PDB Molecule:homeobox protein nkx-3.1;
PDBTitle: solution nmr structure of homeobox domain of homeobox protein nkx-3.12 from homo sapiens, northeast structural genomics consortium target3 hr6470a
|
| 86 | d2cqxa1 |
|
not modelled |
8.8 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 87 | c2ragB_ |
|
not modelled |
8.8 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:dipeptidase;
PDBTitle: crystal structure of aminohydrolase from caulobacter crescentus
|
| 88 | d2cuea1 |
|
not modelled |
8.7 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 89 | c3aogA_ |
|
not modelled |
8.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase;
PDBTitle: crystal structure of glutamate dehydrogenase (gdhb) from thermus2 thermophilus (glu bound form)
|
| 90 | d1au7a1 |
|
not modelled |
8.6 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 91 | c2dmuA_ |
|
not modelled |
8.5 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:homeobox protein goosecoid;
PDBTitle: solution structure of the homeobox domain of homeobox2 protein goosecoid
|
| 92 | d1jgga_ |
|
not modelled |
8.5 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 93 | d2j85a1 |
|
not modelled |
8.5 |
20 |
Fold:STIV B116-like
Superfamily:STIV B116-like
Family:STIV B116-like |
| 94 | c3fdgA_ |
|
not modelled |
8.4 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidase ac. metallo peptidase. merops family m19;
PDBTitle: the crystal structure of the dipeptidase ac, metallo peptidase. merops2 family m19
|
| 95 | d2hddb_ |
|
not modelled |
8.1 |
41 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 96 | d1f43a_ |
|
not modelled |
8.1 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 97 | d1le8a_ |
|
not modelled |
8.0 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 98 | d1ztra1 |
|
not modelled |
7.9 |
41 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 99 | c1ko7B_ |
|
not modelled |
7.9 |
13 |
PDB header:transferase,hydrolase
Chain: B: PDB Molecule:hpr kinase/phosphatase;
PDBTitle: x-ray structure of the hpr kinase/phosphatase from2 staphylococcus xylosus at 1.95 a resolution
|