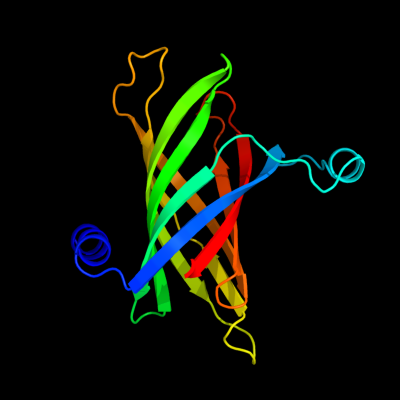
| 1 | d1thqa_
|
|
 |
100.0 |
99 |
Fold:Transmembrane beta-barrels
Superfamily:OMPA-like
Family:Outer membrane enzyme PagP |
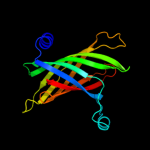
| 2 | d1c8na_
|
|
 |
42.4 |
23 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 3 | c4sbvC_
|
|
 |
42.0 |
27 |
PDB header:virus
Chain: C: PDB Molecule:southern bean mosaic virus coat protein;
PDBTitle: the refinement of southern bean mosaic virus in reciprocal2 space
|
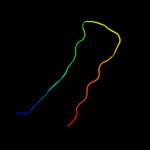
| 4 | d4sbvc_
|
|
 |
42.0 |
27 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 5 | d4sbva_
|
|
 |
40.0 |
27 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
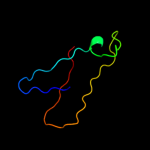
| 6 | d1f2nc_
|
|
 |
39.9 |
21 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 7 | d2vq0a1
|
|
 |
38.2 |
23 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 8 | c1x35C_
|
|
 |
35.0 |
23 |
PDB header:virus
Chain: C: PDB Molecule:coat protein;
PDBTitle: recombinant t=3 capsid of a site specific mutant of semv cp
|
| 9 | d1smva_
|
|
 |
34.3 |
23 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 10 | c2izwC_
|
|
 |
31.7 |
31 |
PDB header:virus
Chain: C: PDB Molecule:ryegrass mottle virus coat protein;
PDBTitle: crystal structure of ryegrass mottle virus
|
| 11 | c1c8nC_
|
|
 |
30.5 |
23 |
PDB header:virus
Chain: C: PDB Molecule:coat protein;
PDBTitle: tobacco necrosis virus
|
| 12 | d1c8nc_
|
|
 |
30.5 |
23 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 13 | c1f2nA_
|
|
 |
30.5 |
15 |
PDB header:virus
Chain: A: PDB Molecule:capsid protein;
PDBTitle: rice yellow mottle virus
|
| 14 | d1f2na_
|
|
 |
30.5 |
15 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 15 | d1ng0c_
|
|
 |
29.1 |
19 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 16 | d1ng0a_
|
|
 |
26.8 |
19 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 17 | c1ng0A_
|
|
 |
26.8 |
19 |
PDB header:virus
Chain: A: PDB Molecule:coat protein;
PDBTitle: the three-dimensional structure of cocksfoot mottle virus2 at 2.7a resolution
|
| 18 | d1smvc_
|
|
 |
24.7 |
23 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 19 | d2tbvc_
|
|
 |
24.3 |
29 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 20 | d2axtz1
|
|
 |
19.7 |
33 |
Fold:Transmembrane helix hairpin
Superfamily:PsbZ-like
Family:PsbZ-like |
| 21 | c2zahC_ |
|
not modelled |
16.6 |
38 |
PDB header:virus
Chain: C: PDB Molecule:coat protein;
PDBTitle: x-ray structure of melon necrotic spot virus
|
| 22 | d1m3ya1 |
|
not modelled |
15.5 |
38 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Group II dsDNA viruses VP
Family:Major capsid protein vp54 |
| 23 | d1opoa_ |
|
not modelled |
12.2 |
23 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Tombusviridae-like VP |
| 24 | c1opoB_ |
|
not modelled |
12.2 |
23 |
PDB header:virus
Chain: B: PDB Molecule:coat protein;
PDBTitle: the structure of carnation mottle virus
|
| 25 | d1bev1_ |
|
not modelled |
12.2 |
23 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 26 | d1cov1_ |
|
not modelled |
10.7 |
22 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 27 | d1igqa_ |
|
not modelled |
9.7 |
26 |
Fold:SH3-like barrel
Superfamily:C-terminal domain of transcriptional repressors
Family:Transcriptional repressor protein KorB |
| 28 | c1m4xC_ |
|
not modelled |
8.3 |
38 |
PDB header:virus
Chain: C: PDB Molecule:pbcv-1 virus capsid;
PDBTitle: pbcv-1 virus capsid, quasi-atomic model
|
| 29 | d1p2za1 |
|
not modelled |
8.3 |
26 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Group II dsDNA viruses VP
Family:Adenovirus hexon |
| 30 | c1j5qB_ |
|
not modelled |
8.3 |
38 |
PDB header:viral protein
Chain: B: PDB Molecule:major capsid protein;
PDBTitle: the structure and evolution of the major capsid protein of a large,2 lipid-containing, dna virus.
|
| 31 | d2d8za2 |
|
not modelled |
8.1 |
56 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 32 | d1igub_ |
|
not modelled |
7.8 |
26 |
Fold:SH3-like barrel
Superfamily:C-terminal domain of transcriptional repressors
Family:Transcriptional repressor protein KorB |
| 33 | d2gr8a1 |
|
not modelled |
7.8 |
29 |
Fold:Pili subunits
Superfamily:Pili subunits
Family:YadA C-terminal domain-like |
| 34 | d1txna_ |
|
not modelled |
7.0 |
29 |
Fold:Coproporphyrinogen III oxidase
Superfamily:Coproporphyrinogen III oxidase
Family:Coproporphyrinogen III oxidase |
| 35 | d1lrwb_ |
|
not modelled |
6.9 |
50 |
Fold:Non-globular all-alpha subunits of globular proteins
Superfamily:Methanol dehydrogenase subunit
Family:Methanol dehydrogenase subunit |
| 36 | c3hdfA_ |
|
not modelled |
6.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysozyme;
PDBTitle: crystal structure of truncated endolysin r21 from phage 21
|
| 37 | c2c1lA_ |
|
not modelled |
6.3 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease;
PDBTitle: structure of the bfii restriction endonuclease
|
| 38 | d1qjpa_ |
|
not modelled |
6.2 |
25 |
Fold:Transmembrane beta-barrels
Superfamily:OMPA-like
Family:Outer membrane protein |
| 39 | c2inyA_ |
|
not modelled |
6.0 |
26 |
PDB header:viral protein
Chain: A: PDB Molecule:hexon protein;
PDBTitle: nanoporous crystals of chicken embryo lethal orphan (celo) adenovirus2 major coat protein, hexon
|
| 40 | c3c5yD_ |
|
not modelled |
5.9 |
43 |
PDB header:isomerase
Chain: D: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
|
| 41 | c2ppwA_ |
|
not modelled |
5.9 |
43 |
PDB header:isomerase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
|
| 42 | d1aa7a_ |
|
not modelled |
5.8 |
20 |
Fold:Influenza virus matrix protein M1
Superfamily:Influenza virus matrix protein M1
Family:Influenza virus matrix protein M1 |
| 43 | c3l4cB_ |
|
not modelled |
5.7 |
28 |
PDB header:cell adhesion, cell invasion, apoptosis
Chain: B: PDB Molecule:dedicator of cytokinesis protein 1;
PDBTitle: structural basis of membrane-targeting by dock180
|
| 44 | d1qj8a_ |
|
not modelled |
5.7 |
20 |
Fold:Transmembrane beta-barrels
Superfamily:OMPA-like
Family:Outer membrane protein |
| 45 | d1t23a_ |
|
not modelled |
5.7 |
46 |
Fold:Chromosomal protein MC1
Superfamily:Chromosomal protein MC1
Family:Chromosomal protein MC1 |
| 46 | c2bviK_ |
|
not modelled |
5.6 |
26 |
PDB header:virus
Chain: K: PDB Molecule:hexon protein;
PDBTitle: the quasi-atomic model of human adenovirus type 52 capsid (part 2)
|
| 47 | d1xmeb2 |
|
not modelled |
5.6 |
50 |
Fold:Transmembrane helix hairpin
Superfamily:Cytochrome c oxidase subunit II-like, transmembrane region
Family:Cytochrome c oxidase subunit II-like, transmembrane region |
| 48 | d1y60a_ |
|
not modelled |
5.6 |
33 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Formaldehyde-activating enzyme, FAE |
| 49 | c3onoA_ |
|
not modelled |
5.5 |
57 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
|
| 50 | c3rpjA_ |
|
not modelled |
5.5 |
25 |
PDB header:transcription regulator
Chain: A: PDB Molecule:curlin genes transcriptional regulator;
PDBTitle: structure of a curlin genes transcriptional regulator protein from2 proteus mirabilis hi4320.
|
| 51 | d2hepa1 |
|
not modelled |
5.4 |
29 |
Fold:Long alpha-hairpin
Superfamily:YnzC-like
Family:YznC-like |
| 52 | c2hepA_ |
|
not modelled |
5.4 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0291 protein ynzc;
PDBTitle: solution nmr structure of the upf0291 protein ynzc from2 bacillus subtilis. northeast structural genomics target3 sr384.
|
| 53 | d1ncqa_ |
|
not modelled |
5.1 |
13 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 54 | c2kebA_ |
|
not modelled |
5.1 |
29 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna polymerase subunit alpha b;
PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
|