| 1 | c2oi6A_ |
|
 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: e. coli glmu- complex with udp-glcnac, coa and glcn-1-po4
|
| 2 | c2v0hA_ |
|
 |
100.0 |
68 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: characterization of substrate binding and catalysis of the2 potential antibacterial target n-acetylglucosamine-1-3 phosphate uridyltransferase (glmu)
|
| 3 | c1hm8A_ |
|
 |
100.0 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine-1-phosphate uridyltransferase;
PDBTitle: crystal structure of s.pneumoniae n-acetylglucosamine-1-phosphate2 uridyltransferase, glmu, bound to acetyl coenzyme a
|
| 4 | c3d98A_ |
|
 |
100.0 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis, ligand-free2 form
|
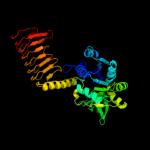
| 5 | c2qkxA_ |
|
 |
100.0 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: n-acetyl glucosamine 1-phosphate uridyltransferase from mycobacterium2 tuberculosis complex with n-acetyl glucosamine 1-phosphate
|
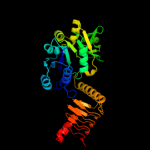
| 6 | c1fwyA_ |
|
 |
100.0 |
90 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of n-acetylglucosamine 1-phosphate2 uridyltransferase bound to udp-glcnac
|
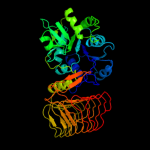
| 7 | c2ggqA_ |
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:401aa long hypothetical glucose-1-phosphate
PDBTitle: complex of hypothetical glucose-1-phosphate thymidylyltransferase from2 sulfolobus tokodaii
|
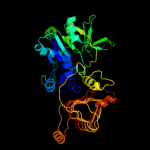
| 8 | c1yp3C_ |
|
 |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:glucose-1-phosphate adenylyltransferase small
PDBTitle: crystal structure of potato tuber adp-glucose2 pyrophosphorylase in complex with atp
|
| 9 | c3brkX_ |
|
 |
100.0 |
15 |
PDB header:transferase
Chain: X: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of adp-glucose pyrophosphorylase from2 agrobacterium tumefaciens
|
| 10 | d2oi6a2 |
|
 |
100.0 |
100 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 11 | c2pa4B_ |
|
 |
100.0 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:utp-glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of udp-glucose pyrophosphorylase from corynebacteria2 glutamicum in complex with magnesium and udp-glucose
|
| 12 | d1lvwa_ |
|
 |
100.0 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 13 | d1fxoa_ |
|
 |
100.0 |
19 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 14 | d1iina_ |
|
 |
100.0 |
21 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 15 | d1g97a2 |
|
 |
100.0 |
41 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 16 | d1h5ra_ |
|
 |
100.0 |
22 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 17 | c2ux8G_ |
|
 |
100.0 |
25 |
PDB header:transferase
Chain: G: PDB Molecule:glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of sphingomonas elodea atcc 31461 glucose-2 1-phosphate uridylyltransferase in complex with glucose-3 1-phosphate.
|
| 18 | d1mc3a_ |
|
 |
100.0 |
17 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 19 | c2e3dB_ |
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:utp--glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of e. coli glucose-1-phosphate2 uridylyltransferase
|
| 20 | c2x5sB_ |
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:mannose-1-phosphate guanylyltransferase;
PDBTitle: crystal structure of t. maritima gdp-mannose2 pyrophosphorylase in apo state.
|
| 21 | c3pnnA_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of a glycosyltransferase from porphyromonas2 gingivalis w83
|
| 22 | c2cu2A_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative mannose-1-phosphate guanylyl transferase;
PDBTitle: crystal structure of mannose-1-phosphate geranyltransferase from2 thermus thermophilus hb8
|
| 23 | c3jukA_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:udp-glucose pyrophosphorylase (galu);
PDBTitle: the crystal structure of udp-glucose pyrophosphorylase complexed with2 udp-glucose
|
| 24 | d1yp2a2 |
|
not modelled |
100.0 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 25 | d1tzfa_ |
|
not modelled |
100.0 |
21 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 26 | c3hl3A_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:glucose-1-phosphate thymidylyltransferase;
PDBTitle: 2.76 angstrom crystal structure of a putative glucose-1-phosphate2 thymidylyltransferase from bacillus anthracis in complex with a3 sucrose.
|
| 27 | d2oi6a1 |
|
not modelled |
100.0 |
100 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 28 | d2cu2a2 |
|
not modelled |
100.0 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:mannose-1-phosphate guanylyl transferase |
| 29 | c1jylC_ |
|
not modelled |
100.0 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:ctp:phosphocholine cytidylytransferase;
PDBTitle: catalytic mechanism of ctp:phosphocholine2 cytidylytransferase from streptococcus pneumoniae (licc)
|
| 30 | c3i3aC_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-
PDBTitle: structural basis for the sugar nucleotide and acyl chain2 selectivity of leptospira interrogans lpxa
|
| 31 | d2jf2a1 |
|
not modelled |
99.9 |
16 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
| 32 | c2iu9C_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine
PDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
|
| 33 | d1jyka_ |
|
not modelled |
99.9 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 34 | c2xwlB_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ispd from mycobacterium smegmatis in complex2 with ctp and mg
|
| 35 | d1j2za_ |
|
not modelled |
99.9 |
17 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
| 36 | d1vica_ |
|
not modelled |
99.9 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 37 | d1g97a1 |
|
not modelled |
99.9 |
40 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 38 | c2qh5B_ |
|
not modelled |
99.9 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:mannose-6-phosphate isomerase;
PDBTitle: crystal structure of mannose-6-phosphate isomerase from helicobacter2 pylori
|
| 39 | c3pmoA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-acyltransferase;
PDBTitle: the structure of lpxd from pseudomonas aeruginosa at 1.3 a resolution
|
| 40 | d1i52a_ |
|
not modelled |
99.9 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 41 | c3r0sA_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: udp-n-acetylglucosamine acyltransferase from campylobacter jejuni
|
| 42 | c2xmhB_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:ctp-inositol-1-phosphate cytidylyltransferase;
PDBTitle: the x-ray structure of ctp:inositol-1-phosphate2 cytidylyltransferase from archaeoglobus fulgidus
|
| 43 | d1vpaa_ |
|
not modelled |
99.9 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 44 | c3eh0C_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-
PDBTitle: crystal structure of lpxd from escherichia coli
|
| 45 | c3oamD_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: crystal structure of cytidylyltransferase from vibrio cholerae
|
| 46 | d1w55a1 |
|
not modelled |
99.9 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 47 | d1h7ea_ |
|
not modelled |
99.9 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 48 | c3polA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: 2.3 angstrom crystal structure of 3-deoxy-manno-octulosonate2 cytidylyltransferase (kdsb) from acinetobacter baumannii.
|
| 49 | c3tqdA_ |
|
not modelled |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: structure of the 3-deoxy-d-manno-octulosonate cytidylyltransferase2 (kdsb) from coxiella burnetii
|
| 50 | d1vh1a_ |
|
not modelled |
99.9 |
20 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 51 | c3fsbB_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:qdtc;
PDBTitle: crystal structure of qdtc, the dtdp-3-amino-3,6-dideoxy-d-2 glucose n-acetyl transferase from thermoanaerobacterium3 thermosaccharolyticum in complex with coa and dtdp-3-amino-4 quinovose
|
| 52 | c2wawA_ |
|
not modelled |
99.9 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:moba relate protein;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0371c2 homolog from mycobacterium sp. strain jc1
|
| 53 | d1vh3a_ |
|
not modelled |
99.9 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 54 | c2wlgA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystallographic analysis of the polysialic acid o-2 acetyltransferase oatwy
|
| 55 | c3jqyB_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystal strucutre of the polysia specific acetyltransferase neuo
|
| 56 | c2y6pC_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: evidence for a two-metal-ion-mechanism in the2 kdo-cytidylyltransferase kdsb
|
| 57 | d1w77a1 |
|
not modelled |
99.9 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 58 | c1w57A_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:ispd/ispf bifunctional enzyme;
PDBTitle: structure of the bifunctional ispdf from campylobacter2 jejuni containing zn
|
| 59 | c3okrA_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
| 60 | c2we9A_ |
|
not modelled |
99.8 |
22 |
PDB header:unknown function
Chain: A: PDB Molecule:moba-related protein;
PDBTitle: crystal structure of rv0371c from mycobacterium2 tuberculosis h37rv
|
| 61 | c3f1cB_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:putative 2-c-methyl-d-erythritol 4-phosphate
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from listeria monocytogenes
|
| 62 | d1eyra_ |
|
not modelled |
99.8 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 63 | c3c8vA_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase;
PDBTitle: crystal structure of putative acetyltransferase (yp_390128.1) from2 desulfovibrio desulfuricans g20 at 2.28 a resolution
|
| 64 | d1krra_ |
|
not modelled |
99.8 |
24 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 65 | d1qwja_ |
|
not modelled |
99.8 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 66 | c1qreA_ |
|
not modelled |
99.8 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: a closer look at the active site of gamma-carbonic anhydrases: high2 resolution crystallographic studies of the carbonic anhydrase from3 methanosarcina thermophila
|
| 67 | d1qrea_ |
|
not modelled |
99.8 |
19 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 68 | d2dpwa1 |
|
not modelled |
99.8 |
20 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:TTHA0179-like |
| 69 | c2ic7A_ |
|
not modelled |
99.8 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:maltose transacetylase;
PDBTitle: crystal structure of maltose transacetylase from2 geobacillus kaustophilus
|
| 70 | c3ectA_ |
|
not modelled |
99.8 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide-repeat containing-acetyltransferase;
PDBTitle: crystal structure of the hexapeptide-repeat containing-2 acetyltransferase vca0836 from vibrio cholerae
|
| 71 | c3srtB_ |
|
not modelled |
99.8 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:maltose o-acetyltransferase;
PDBTitle: the crystal structure of a maltose o-acetyltransferase from2 clostridium difficile 630
|
| 72 | d1mr7a_ |
|
not modelled |
99.8 |
25 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 73 | c3cj8B_ |
|
not modelled |
99.8 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:2,3,4,5-tetrahydropyridine-2-carboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate n-2 succinyltransferase from enterococcus faecalis v583
|
| 74 | c3mqhD_ |
|
not modelled |
99.8 |
24 |
PDB header:transferase
Chain: D: PDB Molecule:lipopolysaccharides biosynthesis acetyltransferase;
PDBTitle: crystal structure of the 3-n-acetyl transferase wlbb from bordetella2 petrii in complex with coa and udp-3-amino-2-acetamido-2,3-dideoxy3 glucuronic acid
|
| 75 | c3fttA_ |
|
not modelled |
99.8 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase sacol2570;
PDBTitle: crystal structure of the galactoside o-acetyltransferase2 from staphylococcus aureus
|
| 76 | d1ocxa_ |
|
not modelled |
99.8 |
25 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 77 | c2px7A_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from thermus thermophilus hb8
|
| 78 | c3eg4A_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase from brucella melitensis3 biovar abortus 2308
|
| 79 | c3rsbB_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:adenosylcobinamide-phosphate guanylyltransferase;
PDBTitle: structure of the archaeal gtp:adocbi-p guanylyltransferase (coby) from2 methanocaldococcus jannaschii
|
| 80 | d1xata_ |
|
not modelled |
99.8 |
31 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 81 | d1e5ka_ |
|
not modelled |
99.8 |
24 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Molybdenum cofactor biosynthesis protein MobA |
| 82 | c2vshB_ |
|
not modelled |
99.8 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate
PDBTitle: synthesis of cdp-activated ribitol for teichoic acid2 precursors in streptococcus pneumoniae
|
| 83 | c3eevC_ |
|
not modelled |
99.7 |
30 |
PDB header:transferase
Chain: C: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: crystal structure of chloramphenicol acetyltransferase vca0300 from2 vibrio cholerae o1 biovar eltor
|
| 84 | d1vgwa_ |
|
not modelled |
99.7 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 85 | c3f1xA_ |
|
not modelled |
99.7 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: three dimensional structure of the serine acetyltransferase from2 bacteroides vulgatus, northeast structural genomics consortium target3 bvr62.
|
| 86 | d1t3da_ |
|
not modelled |
99.7 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
| 87 | d3tdta_ |
|
not modelled |
99.7 |
23 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Tetrahydrodipicolinate-N-succinlytransferase, THDP-succinlytransferase, DapD |
| 88 | c1t3dB_ |
|
not modelled |
99.7 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
|
| 89 | d1ssqa_ |
|
not modelled |
99.7 |
24 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
| 90 | c3ngwA_ |
|
not modelled |
99.7 |
20 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin-guanine dinucleotide biosynthesis protein a
PDBTitle: crystal structure of molybdopterin-guanine dinucleotide biosynthesis2 protein a from archaeoglobus fulgidus, northeast structural genomics3 consortium target gr189
|
| 91 | c3mc4A_ |
|
not modelled |
99.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:ww/rsp5/wwp domain:bacterial transferase
PDBTitle: crystal structure of ww/rsp5/wwp domain: bacterial2 transferase hexapeptide repeat: serine o-acetyltransferase3 from brucella melitensis
|
| 92 | d3bswa1 |
|
not modelled |
99.7 |
15 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:PglD-like |
| 93 | c3q1xA_ |
|
not modelled |
99.7 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of entamoeba histolytica serine acetyltransferase 12 in complex with l-serine
|
| 94 | c3r3rA_ |
|
not modelled |
99.7 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:ferripyochelin binding protein;
PDBTitle: structure of the yrda ferripyochelin binding protein from salmonella2 enterica
|
| 95 | c2e8bA_ |
|
not modelled |
99.7 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable molybdopterin-guanine dinucleotide biosynthesis
PDBTitle: crystal structure of the putative protein (aq1419) from aquifex2 aeolicus vf5
|
| 96 | c3r1wA_ |
|
not modelled |
99.7 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of a carbonic anhydrase from a crude oil degrading2 psychrophilic library
|
| 97 | d1v3wa_ |
|
not modelled |
99.7 |
21 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 98 | d2f9ca1 |
|
not modelled |
99.7 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:YdcK-like |
| 99 | d1xhda_ |
|
not modelled |
99.6 |
26 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 100 | c3ixcA_ |
|
not modelled |
99.6 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide transferase family protein;
PDBTitle: crystal structure of hexapeptide transferase family protein from2 anaplasma phagocytophilum
|
| 101 | c3fsyC_ |
|
not modelled |
99.6 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:tetrahydrodipicolinate n-succinyltransferase;
PDBTitle: structure of tetrahydrodipicolinate n-succinyltransferase2 (rv1201c;dapd) in complex with succinyl-coa from mycobacterium3 tuberculosis
|
| 102 | c3okrC_ |
|
not modelled |
99.6 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
| 103 | c3kwdA_ |
|
not modelled |
99.5 |
18 |
PDB header:lyase, protein binding, photosynthesis
Chain: A: PDB Molecule:carbon dioxide concentrating mechanism protein;
PDBTitle: inactive truncation of the beta-carboxysomal gamma-carbonic anhydrase,2 ccmm, form 1
|
| 104 | c3d5nB_ |
|
not modelled |
99.4 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:q97w15_sulso;
PDBTitle: crystal structure of the q97w15_sulso protein from2 sulfolobus solfataricus. nesg target ssr125.
|
| 105 | d1yp2a1 |
|
not modelled |
99.3 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 106 | c2rijA_ |
|
not modelled |
99.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:putative 2,3,4,5-tetrahydropyridine-2-carboxylate n-
PDBTitle: crystal structure of a putative 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase (cj1605c, dapd) from campylobacter3 jejuni at 1.90 a resolution
|
| 107 | d1fxja1 |
|
not modelled |
99.0 |
24 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 108 | c2i5kB_ |
|
not modelled |
98.8 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:utp--glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of ugp1p
|
| 109 | c3oc9A_ |
|
not modelled |
98.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of putative udp-n-acetylglucosamine2 pyrophosphorylase from entamoeba histolytica
|
| 110 | c2yqsA_ |
|
not modelled |
98.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of uridine-diphospho-n-acetylglucosamine2 pyrophosphorylase from candida albicans, in the product-binding form
|
| 111 | d1vm8a_ |
|
not modelled |
98.6 |
20 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 112 | d1jv1a_ |
|
not modelled |
98.6 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 113 | d2icya2 |
|
not modelled |
98.5 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 114 | c3gueB_ |
|
not modelled |
98.1 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:utp-glucose-1-phosphate uridylyltransferase 2;
PDBTitle: crystal structure of udp-glucose phosphorylase from trypanosoma2 brucei, (tb10.389.0330)
|
| 115 | c2q4jB_ |
|
not modelled |
98.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:probable utp-glucose-1-phosphate uridylyltransferase 2;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at3g03250, a putative udp-glucose3 pyrophosphorylase
|
| 116 | c3ogzA_ |
|
not modelled |
98.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:udp-sugar pyrophosphorylase;
PDBTitle: protein structure of usp from l. major in apo-form
|
| 117 | c2oefA_ |
|
not modelled |
97.4 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:utp-glucose-1-phosphate uridylyltransferase 2,
PDBTitle: open and closed structures of the udp-glucose2 pyrophosphorylase from leishmania major
|
| 118 | c3cgxA_ |
|
not modelled |
94.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:putative nucleotide-diphospho-sugar transferase;
PDBTitle: crystal structure of putative nucleotide-diphospho-sugar transferase2 (yp_389115.1) from desulfovibrio desulfuricans g20 at 1.90 a3 resolution
|
| 119 | d1omza_ |
|
not modelled |
92.0 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Exostosin |
| 120 | c1omxB_ |
|
not modelled |
90.4 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:alpha-1,4-n-acetylhexosaminyltransferase extl2;
PDBTitle: crystal structure of mouse alpha-1,4-n-2 acetylhexosaminyltransferase (extl2)
|

