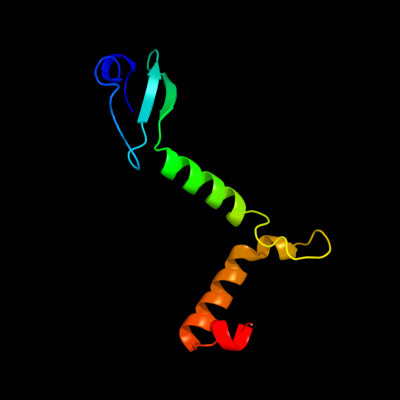
1 c3g5oA_
97.9
17
PDB header: toxin/antitoxinChain: A: PDB Molecule: uncharacterized protein rv2865;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
2 c3hs2H_
97.8
22
PDB header: antitoxinChain: H: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd truncated to residue 57 in an orthorhombic2 space group
3 d2a6qa1
97.4
20
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like4 c2odkD_
97.2
11
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: putative prevent-host-death protein from nitrosomonas europaea
5 d2odka1
97.2
11
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like6 d2a6qb1
97.2
20
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like7 c3hryA_
97.1
20
PDB header: antitoxinChain: A: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd in a trigonal space group and partially2 disordered
8 c1f3mB_
22.2
27
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase pak-alpha;PDBTitle: crystal structure of human serine/threonine kinase pak1
9 c2k9iB_
17.2
17
PDB header: dna binding proteinChain: B: PDB Molecule: plasmid prn1, complete sequence;PDBTitle: nmr structure of plasmid copy control protein orf56 from2 sulfolobus islandicus
10 c3nutC_
16.3
25
PDB header: transferaseChain: C: PDB Molecule: precorrin-3 methylase;PDBTitle: crystal structure of the methyltransferase cobj
11 d1zcea1
14.6
15
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like12 c3owvA_
14.2
7
PDB header: hydrolaseChain: A: PDB Molecule: dna-entry nuclease;PDBTitle: structural insights into catalytic and substrate binding mechanisms of2 the strategic enda nuclease from streptococcus pneumoniae
13 d3kvta_
12.4
10
Fold: POZ domainSuperfamily: POZ domainFamily: Tetramerization domain of potassium channels14 d2gbsa1
11.7
18
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like15 c3trrA_
11.4
13
PDB header: isomeraseChain: A: PDB Molecule: probable enoyl-coa hydratase/isomerase;PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
16 c3swxB_
11.2
23
PDB header: isomeraseChain: B: PDB Molecule: probable enoyl-coa hydratase/isomerase;PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
17 c3he2C_
11.1
16
PDB header: lyaseChain: C: PDB Molecule: enoyl-coa hydratase echa6;PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium2 tuberculosis
18 c3gqcB_
9.8
41
PDB header: transferase/dnaChain: B: PDB Molecule: dna repair protein rev1;PDBTitle: structure of human rev1-dna-dntp ternary complex
19 c2o18A_
9.4
7
PDB header: lipid binding proteinChain: A: PDB Molecule: thiamine biosynthesis lipoprotein apbe;PDBTitle: crystal structure of a thiamine biosynthesis lipoprotein2 apbe, northeast strcutural genomics target er559
20 c3a5zF_
9.4
18
PDB header: ligaseChain: F: PDB Molecule: elongation factor p;PDBTitle: crystal structure of escherichia coli genx in complex with elongation2 factor p
21 c1vdbA_
not modelled
9.3
41
PDB header: transcriptionChain: A: PDB Molecule: fibroin-modulator-binding-protein-1;PDBTitle: nmr structure of fmbp-1 tandem repeat 1 in 30%(v/v) tfe2 solution
22 c3eopB_
not modelled
9.1
16
PDB header: unknown functionChain: B: PDB Molecule: thymocyte nuclear protein 1;PDBTitle: crystal structure of the duf55 domain of human thymocyte nuclear2 protein 1
23 d2ar1a1
not modelled
8.7
25
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like24 c2e0kA_
not modelled
8.7
19
PDB header: transferaseChain: A: PDB Molecule: precorrin-2 c20-methyltransferase;PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
25 c2wjeA_
not modelled
8.4
11
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase cpsb;PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
26 c3d89A_
not modelled
8.2
13
PDB header: electron transportChain: A: PDB Molecule: rieske domain-containing protein;PDBTitle: crystal structure of a soluble rieske ferredoxin from mus musculus
27 c2x5fB_
not modelled
8.1
17
PDB header: transferaseChain: B: PDB Molecule: aspartate_tyrosine_phenylalanine pyridoxal-5'PDBTitle: crystal structure of the methicillin-resistant2 staphylococcus aureus sar2028, an3 aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate4 dependent aminotransferase
28 d2evea1
not modelled
8.0
10
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like29 c2fllA_
not modelled
7.6
26
PDB header: replication/dnaChain: A: PDB Molecule: dna polymerase iota;PDBTitle: ternary complex of human dna polymerase iota with dna and dttp
30 d2o16a3
not modelled
7.2
19
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair31 d1uiya_
not modelled
7.2
25
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Crotonase-like32 c2ngrB_
not modelled
7.2
15
PDB header: hydrolaseChain: B: PDB Molecule: protein (gtpase activating protein (rhg));PDBTitle: transition state complex for gtp hydrolysis by cdc42:2 comparisons of the high resolution structures for cdc423 bound to the active and catalytically compromised forms of4 the cdc42-gap.
33 c2kc8B_
not modelled
7.0
29
PDB header: toxin/toxin repressorChain: B: PDB Molecule: antitoxin relb;PDBTitle: structure of e. coli toxin rele (r81a/r83a) mutant in2 complex with antitoxin relbc (k47-l79) peptide
34 c1t3nB_
not modelled
7.0
26
PDB header: replication/dnaChain: B: PDB Molecule: polymerase (dna directed) iota;PDBTitle: structure of the catalytic core of dna polymerase iota in2 complex with dna and dttp
35 d1qkia1
not modelled
6.9
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain36 c2fwtA_
not modelled
6.7
41
PDB header: electron transportChain: A: PDB Molecule: dhc, diheme cytochrome c;PDBTitle: crystal structure of dhc purified from rhodobacter2 sphaeroides
37 c2bb3B_
not modelled
6.7
31
PDB header: transferaseChain: B: PDB Molecule: cobalamin biosynthesis precorrin-6y methylase (cbie);PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
38 d1uxda_
not modelled
6.6
24
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator39 c3lkeA_
not modelled
6.5
25
PDB header: lyaseChain: A: PDB Molecule: enoyl-coa hydratase;PDBTitle: crystal structure of enoyl-coa hydratase from bacillus2 halodurans
40 d1t94a2
not modelled
6.3
30
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: Lesion bypass DNA polymerase (Y-family), catalytic domain41 c1s97D_
not modelled
6.3
15
PDB header: transferase/dnaChain: D: PDB Molecule: dna polymerase iv;PDBTitle: dpo4 with gt mismatch
42 c2i2rK_
not modelled
6.2
9
PDB header: transport proteinChain: K: PDB Molecule: potassium voltage-gated channel subfamily d member 3;PDBTitle: crystal structure of the kchip1/kv4.3 t1 complex
43 d1vrma1
not modelled
5.9
12
Fold: T-foldSuperfamily: ApbE-likeFamily: ApbE-like44 c2l7kA_
not modelled
5.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein cd1104.2 from clostridium difficile,2 northeast structural genomics consortium target cfr130
45 c2q35A_
not modelled
5.9
19
PDB header: lyaseChain: A: PDB Molecule: curf;PDBTitle: crystal structure of the y82f variant of ech2 decarboxylase domain of2 curf from lyngbya majuscula
46 c2ahpB_
not modelled
5.9
20
PDB header: de novo proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper, mutation of lys15 to epsilon-azido-lys
47 d1exbe_
not modelled
5.8
9
Fold: POZ domainSuperfamily: POZ domainFamily: Tetramerization domain of potassium channels48 c1untA_
not modelled
5.8
38
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
49 d1xn7a_
not modelled
5.7
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Hypothetical protein YhgG50 c2ahpA_
not modelled
5.6
20
PDB header: de novo proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper, mutation of lys15 to epsilon-azido-lys
51 c3eevC_
not modelled
5.6
23
PDB header: transferaseChain: C: PDB Molecule: chloramphenicol acetyltransferase;PDBTitle: crystal structure of chloramphenicol acetyltransferase vca0300 from2 vibrio cholerae o1 biovar eltor
52 c1ciyA_
not modelled
5.5
14
PDB header: toxinChain: A: PDB Molecule: cryia(a);PDBTitle: insecticidal toxin: structure and channel formation
53 c2oh2B_
not modelled
5.4
39
PDB header: transferase/dnaChain: B: PDB Molecule: dna polymerase kappa;PDBTitle: ternary complex of human dna polymerase
54 d1t1da_
not modelled
5.4
9
Fold: POZ domainSuperfamily: POZ domainFamily: Tetramerization domain of potassium channels55 c1untB_
not modelled
5.3
38
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
56 d1zeta2
not modelled
5.3
26
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: Lesion bypass DNA polymerase (Y-family), catalytic domain57 d1xata_
not modelled
5.3
13
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like58 c1t94B_
not modelled
5.3
39
PDB header: replicationChain: B: PDB Molecule: polymerase (dna directed) kappa;PDBTitle: crystal structure of the catalytic core of human dna2 polymerase kappa