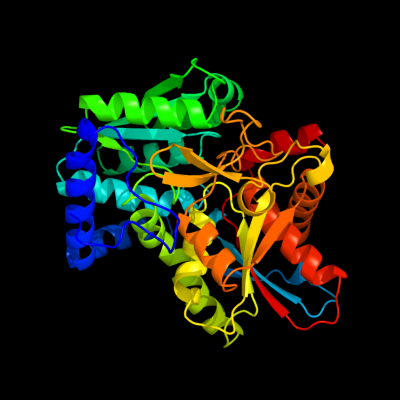
| 1 | d1qopb_
|
|
 |
100.0 |
97 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
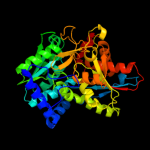
| 2 | c1x1qA_
|
|
 |
100.0 |
54 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
|
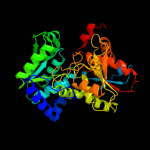
| 3 | d1v8za1
|
|
 |
100.0 |
59 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
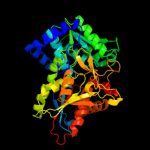
| 4 | c2o2jA_
|
|
 |
100.0 |
59 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: mycobacterium tuberculosis tryptophan synthase beta chain2 dimer (apoform)
|
| 5 | d1wkva1
|
|
 |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 6 | c1tdjA_
|
|
 |
100.0 |
21 |
PDB header:allostery
Chain: A: PDB Molecule:biosynthetic threonine deaminase;
PDBTitle: threonine deaminase (biosynthetic) from e. coli
|
| 7 | c3iauA_
|
|
 |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:threonine deaminase;
PDBTitle: the structure of the processed form of threonine deaminase isoform 22 from solanum lycopersicum
|
| 8 | c3pc3A_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:cg1753, isoform a;
PDBTitle: full length structure of cystathionine beta-synthase from drosophila2 in complex with aminoacrylate
|
| 9 | d1v71a1
|
|
 |
100.0 |
22 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 10 | d1jbqa_
|
|
 |
100.0 |
22 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 11 | c1jbqD_
|
|
 |
100.0 |
22 |
PDB header:lyase
Chain: D: PDB Molecule:cystathionine beta-synthase;
PDBTitle: structure of human cystathionine beta-synthase: a unique pyridoxal 5'-2 phosphate dependent hemeprotein
|
| 12 | c2gn0A_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:threonine dehydratase catabolic;
PDBTitle: crystal structure of dimeric biodegradative threonine deaminase (tdcb)2 from salmonella typhimurium at 1.7 a resolution (triclinic form with3 one complete subunit built in alternate conformation)
|
| 13 | d1z7wa1
|
|
 |
100.0 |
22 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 14 | d1pwha_
|
|
 |
100.0 |
22 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 15 | d1tdja1
|
|
 |
100.0 |
21 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 16 | c3r0zA_
|
|
 |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:d-serine dehydratase;
PDBTitle: crystal structure of apo d-serine deaminase from salmonella2 typhimurium
|
| 17 | c3l6cA_
|
|
 |
100.0 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:serine racemase;
PDBTitle: x-ray crystal structure of rat serine racemase in complex with2 malonate a potent inhibitor
|
| 18 | d1ve5a1
|
|
 |
100.0 |
23 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 19 | c1p5jA_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:l-serine dehydratase;
PDBTitle: crystal structure analysis of human serine dehydratase
|
| 20 | d1p5ja_
|
|
 |
100.0 |
21 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 21 | d1ve1a1 |
|
not modelled |
100.0 |
23 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 22 | c2pqmA_ |
|
not modelled |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:cysteine synthase;
PDBTitle: crystal structure of cysteine synthase (oass) from entamoeba2 histolytica at 1.86 a resolution
|
| 23 | d2bhsa1 |
|
not modelled |
100.0 |
24 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 24 | c3dwgA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase b;
PDBTitle: crystal structure of a sulfur carrier protein complex found in the2 cysteine biosynthetic pathway of mycobacterium tuberculosis
|
| 25 | d1y7la1 |
|
not modelled |
100.0 |
19 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 26 | c2q3bA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase a;
PDBTitle: 1.8 a resolution crystal structure of o-acetylserine sulfhydrylase2 (oass) holoenzyme from mycobacterium tuberculosis
|
| 27 | c2d1fA_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:threonine synthase;
PDBTitle: structure of mycobacterium tuberculosis threonine synthase
|
| 28 | d1v7ca_ |
|
not modelled |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 29 | c2zsjB_ |
|
not modelled |
100.0 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:threonine synthase;
PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
|
| 30 | d1o58a_ |
|
not modelled |
100.0 |
24 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 31 | c2rkbE_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: E: PDB Molecule:serine dehydratase-like;
PDBTitle: serine dehydratase like-1 from human cancer cells
|
| 32 | c2eguA_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase;
PDBTitle: crystal structure of o-acetylserine sulfhydrase from geobacillus2 kaustophilus hta426
|
| 33 | d1fcja_ |
|
not modelled |
100.0 |
22 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 34 | d1f2da_ |
|
not modelled |
100.0 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 35 | d1j0aa_ |
|
not modelled |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 36 | d1e5xa_ |
|
not modelled |
100.0 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 37 | d1tyza_ |
|
not modelled |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 38 | d1vb3a1 |
|
not modelled |
100.0 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 39 | d1kl7a_ |
|
not modelled |
100.0 |
12 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 40 | d1vp8a_ |
|
not modelled |
95.2 |
16 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 41 | c3iupB_ |
|
not modelled |
92.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadph:quinone oxidoreductase;
PDBTitle: crystal structure of putative nadph:quinone oxidoreductase2 (yp_296108.1) from ralstonia eutropha jmp134 at 1.70 a resolution
|
| 42 | d1c1da1 |
|
not modelled |
86.7 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 43 | c3o93A_ |
|
not modelled |
85.7 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: high resolution crystal structures of streptococcus pneumoniae2 nicotinamidase with trapped intermediates provide insights into3 catalytic mechanism and inhibition by aldehydes
|
| 44 | d1bg6a2 |
|
not modelled |
85.5 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 45 | d1hyua1 |
|
not modelled |
84.4 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 46 | c3krtC_ |
|
not modelled |
84.0 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:crotonyl coa reductase;
PDBTitle: crystal structure of putative crotonyl coa reductase from streptomyces2 coelicolor a3(2)
|
| 47 | d1o8ca2 |
|
not modelled |
84.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 48 | d1l7da1 |
|
not modelled |
83.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 49 | d1nbaa_ |
|
not modelled |
83.4 |
12 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 50 | c3d4oA_ |
|
not modelled |
80.6 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 51 | c3k30B_ |
|
not modelled |
80.3 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
| 52 | d1t57a_ |
|
not modelled |
80.1 |
10 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 53 | c2rghA_ |
|
not modelled |
77.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
| 54 | d1fl2a1 |
|
not modelled |
76.0 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 55 | d1im5a_ |
|
not modelled |
74.1 |
15 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 56 | c1boiA_ |
|
not modelled |
73.3 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:rhodanese;
PDBTitle: n-terminally truncated rhodanese
|
| 57 | c3gbcA_ |
|
not modelled |
73.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrazinamidase/nicotinamidas pnca;
PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
|
| 58 | c2bruB_ |
|
not modelled |
70.6 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase subunit alpha;
PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
|
| 59 | c3oqpB_ |
|
not modelled |
70.4 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 60 | d1trba1 |
|
not modelled |
70.1 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 61 | d1vdca1 |
|
not modelled |
69.1 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 62 | c3gmbB_ |
|
not modelled |
69.1 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic2 acid oxygenase
|
| 63 | c1gl9B_ |
|
not modelled |
68.9 |
13 |
PDB header:topoisomerase
Chain: B: PDB Molecule:reverse gyrase;
PDBTitle: archaeoglobus fulgidus reverse gyrase complexed with adpnp
|
| 64 | c2ywlA_ |
|
not modelled |
68.4 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase related protein;
PDBTitle: crystal structure of thioredoxin reductase-related protein ttha03702 from thermus thermophilus hb8
|
| 65 | c2qx7A_ |
|
not modelled |
68.3 |
18 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
| 66 | d1seza1 |
|
not modelled |
68.1 |
28 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 67 | c3da1A_ |
|
not modelled |
68.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: x-ray structure of the glycerol-3-phosphate dehydrogenase2 from bacillus halodurans complexed with fad. northeast3 structural genomics consortium target bhr167.
|
| 68 | d2i0za1 |
|
not modelled |
67.9 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
| 69 | c3d3kD_ |
|
not modelled |
67.5 |
9 |
PDB header:protein binding
Chain: D: PDB Molecule:enhancer of mrna-decapping protein 3;
PDBTitle: crystal structure of human edc3p
|
| 70 | d1mv8a2 |
|
not modelled |
67.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 71 | d1tt7a2 |
|
not modelled |
66.3 |
8 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 72 | d1xgka_ |
|
not modelled |
66.1 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 73 | d3lada1 |
|
not modelled |
65.5 |
41 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 74 | c3oqpA_ |
|
not modelled |
65.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 75 | d1o89a2 |
|
not modelled |
64.7 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 76 | d1ml4a2 |
|
not modelled |
64.3 |
12 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 77 | d1dxla1 |
|
not modelled |
63.9 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 78 | c3orqA_ |
|
not modelled |
63.2 |
23 |
PDB header:ligase,biosynthetic protein
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide synthetase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
|
| 79 | c3hu5B_ |
|
not modelled |
63.0 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein from desulfovibrio2 vulgaris subsp. vulgaris str. hildenborough
|
| 80 | d2cula1 |
|
not modelled |
62.5 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GidA-like |
| 81 | d1ebda1 |
|
not modelled |
62.4 |
37 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 82 | d1lvla1 |
|
not modelled |
62.3 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 83 | d3lada2 |
|
not modelled |
61.4 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 84 | d1v59a2 |
|
not modelled |
61.3 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 85 | c3eywA_ |
|
not modelled |
60.9 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 86 | c3ot4F_ |
|
not modelled |
60.8 |
19 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative isochorismatase;
PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
|
| 87 | c2rgoA_ |
|
not modelled |
60.7 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
| 88 | c1bg6A_ |
|
not modelled |
60.5 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;
PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
|
| 89 | d1ryia1 |
|
not modelled |
60.5 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 90 | c3allA_ |
|
not modelled |
59.6 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
|
| 91 | c3p2yA_ |
|
not modelled |
59.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alanine dehydrogenase/pyridine nucleotide transhydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
|
| 92 | d2gf3a1 |
|
not modelled |
58.7 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 93 | d1onfa2 |
|
not modelled |
58.5 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 94 | c1djnB_ |
|
not modelled |
58.3 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trimethylamine dehydrogenase;
PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
|
| 95 | c2vhyB_ |
|
not modelled |
58.2 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of apo l-alanine dehydrogenase from2 mycobacterium tuberculosis
|
| 96 | c2hk8B_ |
|
not modelled |
57.7 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
|
| 97 | c3c1oA_ |
|
not modelled |
57.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol synthase;
PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
|
| 98 | c3eplA_ |
|
not modelled |
56.9 |
15 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna isopentenyltransferase;
PDBTitle: crystallographic snapshots of eukaryotic2 dimethylallyltransferase acting on trna: insight into trna3 recognition and reaction mechanism
|
| 99 | d1neka2 |
|
not modelled |
56.5 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 100 | c2bryA_ |
|
not modelled |
56.5 |
26 |
PDB header:transport
Chain: A: PDB Molecule:nedd9 interacting protein with calponin homology
PDBTitle: crystal structure of the native monooxygenase domain of2 mical at 1.45 a resolution
|
| 101 | c3k96B_ |
|
not modelled |
56.0 |
37 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
| 102 | c3g05B_ |
|
not modelled |
55.2 |
26 |
PDB header:rna binding protein
Chain: B: PDB Molecule:trna uridine 5-carboxymethylaminomethyl modification enzyme
PDBTitle: crystal structure of n-terminal domain (2-550) of e.coli mnmg
|
| 103 | d2iida1 |
|
not modelled |
55.0 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 104 | c3fk4A_ |
|
not modelled |
55.0 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:rubisco-like protein;
PDBTitle: crystal structure of rubisco-like protein from bacillus2 cereus atcc 14579
|
| 105 | d1qyda_ |
|
not modelled |
54.9 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 106 | d1ojta1 |
|
not modelled |
54.7 |
37 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 107 | c2xdoC_ |
|
not modelled |
54.3 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:tetx2 protein;
PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
|
| 108 | c3f8rD_ |
|
not modelled |
53.8 |
28 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:thioredoxin reductase (trxb-3);
PDBTitle: crystal structure of sulfolobus solfataricus thioredoxin2 reductase b3 in complex with two nadp molecules
|
| 109 | d1lpfa1 |
|
not modelled |
53.7 |
37 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 110 | c2ze5A_ |
|
not modelled |
53.6 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:isopentenyl transferase;
PDBTitle: crystal structure of adenosine phosphate-isopentenyltransferase
|
| 111 | c3cumA_ |
|
not modelled |
53.3 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
| 112 | c3atrA_ |
|
not modelled |
53.2 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:conserved archaeal protein;
PDBTitle: geranylgeranyl reductase (ggr) from sulfolobus acidocaldarius co-2 crystallized with its ligand
|
| 113 | c1hyuA_ |
|
not modelled |
53.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: crystal structure of intact ahpf
|
| 114 | c1pjcA_ |
|
not modelled |
52.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (l-alanine dehydrogenase);
PDBTitle: l-alanine dehydrogenase complexed with nad
|
| 115 | d2bs2a2 |
|
not modelled |
52.8 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 116 | d1kjqa2 |
|
not modelled |
52.8 |
22 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 117 | c2vdcI_ |
|
not modelled |
52.5 |
28 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glutamate synthase [nadph] small chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 118 | c3d3jA_ |
|
not modelled |
51.9 |
9 |
PDB header:protein binding
Chain: A: PDB Molecule:enhancer of mrna-decapping protein 3;
PDBTitle: crystal structure of human edc3p
|
| 119 | c3cgvA_ |
|
not modelled |
51.8 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:geranylgeranyl reductase related protein;
PDBTitle: crystal structure of geranylgeranyl bacteriochlorophyll reductase-like2 fixc homolog (np_393992.1) from thermoplasma acidophilum at 1.60 a3 resolution
|
| 120 | c2gf2B_ |
|
not modelled |
51.6 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of human hydroxyisobutyrate dehydrogenase
|