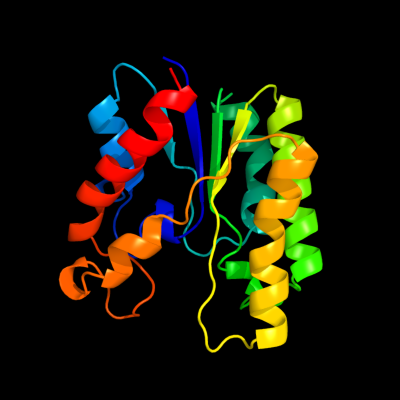
| 1 | d1jrla_
|
|
 |
100.0 |
99 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:TAP-like |
| 2 | c3hp4A_
|
|
 |
100.0 |
44 |
PDB header:hydrolase
Chain: A: PDB Molecule:gdsl-esterase;
PDBTitle: crystal structure of psychrotrophic esterase esta from2 pseudoalteromonas sp. 643a inhibited by monoethylphosphonate
|
| 3 | c2o14A_
|
|
 |
100.0 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yxim;
PDBTitle: x-ray crystal structure of protein yxim_bacsu from bacillus2 subtilis. northeast structural genomics consortium target3 sr595
|
| 4 | c3milA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:isoamyl acetate-hydrolyzing esterase;
PDBTitle: crystal structure of isoamyl acetate-hydrolyzing esterase from2 saccharomyces cerevisiae
|
| 5 | c3p94A_
|
|
 |
100.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:gdsl-like lipase;
PDBTitle: crystal structure of a gdsl-like lipase (bdi_0976) from2 parabacteroides distasonis atcc 8503 at 1.93 a resolution
|
| 6 | c3rjtA_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipolytic protein g-d-s-l family;
PDBTitle: crystal structure of lipolytic protein g-d-s-l family from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
|
| 7 | d1yzfa1
|
|
 |
100.0 |
18 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:TAP-like |
| 8 | c2vptA_
|
|
 |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipolytic enzyme;
PDBTitle: clostridium thermocellum family 3 carbohydrate esterase
|
| 9 | d3bzwa1
|
|
 |
100.0 |
17 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:BT2961-like |
| 10 | c3bzwA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative lipase;
PDBTitle: crystal structure of a putative lipase from bacteroides2 thetaiotaomicron
|
| 11 | c2waoA_
|
|
 |
99.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase e;
PDBTitle: structure of a family two carbohydrate esterase from2 clostridium thermocellum in complex with cellohexaose
|
| 12 | d1k7ca_
|
|
 |
99.9 |
18 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Rhamnogalacturonan acetylesterase |
| 13 | c2waaA_
|
|
 |
99.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:xylan esterase, putative, axe2c;
PDBTitle: structure of a family two carbohydrate esterase from2 cellvibrio japonicus
|
| 14 | d2o14a2
|
|
 |
99.9 |
24 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:YxiM C-terminal domain-like |
| 15 | d3dc7a1
|
|
 |
99.9 |
19 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:BT2961-like |
| 16 | c3dc7B_
|
|
 |
99.9 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein lp_3323;
PDBTitle: crystal structure of the protein q88sr8 from lactobacillus plantarum.2 northeast structural genomics consortium target lpr109.
|
| 17 | d1vjga_
|
|
 |
99.9 |
20 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Hypothetical protein alr1529 |
| 18 | d1es9a_
|
|
 |
99.9 |
20 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Acetylhydrolase |
| 19 | d1fxwf_
|
|
 |
99.9 |
19 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Acetylhydrolase |
| 20 | c2q0qC_
|
|
 |
99.9 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:aryl esterase;
PDBTitle: structure of the native m. smegmatis aryl esterase
|
| 21 | c3dciB_ |
|
not modelled |
99.9 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:arylesterase;
PDBTitle: the structure of a putative arylesterase from agrobacterium2 tumefaciens str. c58
|
| 22 | d2hsja1 |
|
not modelled |
99.9 |
22 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Acetylhydrolase |
| 23 | c2w9xA_ |
|
not modelled |
99.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative acetyl xylan esterase;
PDBTitle: the active site of a carbohydrate esterase displays2 divergent3 catalytic and non-catalytic binding functions
|
| 24 | d1esca_ |
|
not modelled |
99.9 |
21 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Esterase |
| 25 | c3kvnA_ |
|
not modelled |
98.7 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:esterase esta;
PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
|
| 26 | d2apja1 |
|
not modelled |
97.6 |
11 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Putative acetylxylan esterase-like |
| 27 | c3bmaC_ |
|
not modelled |
97.0 |
9 |
PDB header:ligase
Chain: C: PDB Molecule:d-alanyl-lipoteichoic acid synthetase;
PDBTitle: crystal structure of d-alanyl-lipoteichoic acid synthetase from2 streptococcus pneumoniae r6
|
| 28 | c3pt5A_ |
|
not modelled |
96.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:nans (yjhs), a 9-o-acetyl n-acetylneuraminic acid esterase;
PDBTitle: crystal structure of nans
|
| 29 | d1zmba1 |
|
not modelled |
96.3 |
14 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Putative acetylxylan esterase-like |
| 30 | c3nvbA_ |
|
not modelled |
88.0 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of n-terminal part of the protein bf1531 from2 bacteroides fragilis containing phosphatase domain complexed with mg3 and tungstate
|
| 31 | d1jlja_ |
|
not modelled |
87.7 |
14 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 32 | d2f7wa1 |
|
not modelled |
85.9 |
14 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 33 | c2pjkA_ |
|
not modelled |
84.9 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:178aa long hypothetical molybdenum cofactor
PDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
|
| 34 | d1mkza_ |
|
not modelled |
82.9 |
15 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 35 | c3q9cF_ |
|
not modelled |
81.5 |
12 |
PDB header:hydrolase
Chain: F: PDB Molecule:acetylpolyamine amidohydrolase;
PDBTitle: crystal structure of h159a apah complexed with n8-acetylspermidine
|
| 36 | c3menC_ |
|
not modelled |
80.4 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:acetylpolyamine aminohydrolase;
PDBTitle: crystal structure of acetylpolyamine aminohydrolase from burkholderia2 pseudomallei, iodide soak
|
| 37 | c1vcnA_ |
|
not modelled |
77.7 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:ctp synthetase;
PDBTitle: crystal structure of t.th. hb8 ctp synthetase complex with sulfate2 anion
|
| 38 | d1uuya_ |
|
not modelled |
77.4 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 39 | d1vcoa2 |
|
not modelled |
77.2 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 40 | c2is8A_ |
|
not modelled |
76.6 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
| 41 | d1y5ea1 |
|
not modelled |
72.4 |
14 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 42 | c3maxB_ |
|
not modelled |
71.7 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:histone deacetylase 2;
PDBTitle: crystal structure of human hdac2 complexed with an n-(2-aminophenyl)2 benzamide
|
| 43 | d1t64a_ |
|
not modelled |
70.8 |
18 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Histone deacetylase, HDAC |
| 44 | c3ew8A_ |
|
not modelled |
70.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:histone deacetylase 8;
PDBTitle: crystal structure analysis of human hdac8 d101l variant
|
| 45 | d2csua1 |
|
not modelled |
69.8 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 46 | c3nvaB_ |
|
not modelled |
69.5 |
12 |
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: dimeric form of ctp synthase from sulfolobus solfataricus
|
| 47 | d1c3pa_ |
|
not modelled |
55.9 |
16 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Histone deacetylase, HDAC |
| 48 | c3qi7A_ |
|
not modelled |
55.9 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of a putative transcriptional regulator2 (yp_001089212.1) from clostridium difficile 630 at 1.86 a resolution
|
| 49 | c2bg5C_ |
|
not modelled |
47.2 |
8 |
PDB header:transferase
Chain: C: PDB Molecule:phosphoenolpyruvate-protein kinase;
PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme2 i-domain from the thermoanaerobacter tengcongensis pep:3 sugar phosphotransferase system (pts)
|
| 50 | c2g4rB_ |
|
not modelled |
44.1 |
15 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis mog protein;
PDBTitle: anomalous substructure of moga
|
| 51 | c1y8aA_ |
|
not modelled |
40.7 |
45 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein af1437;
PDBTitle: structure of gene product af1437 from archaeoglobus fulgidus
|
| 52 | d2g2ca1 |
|
not modelled |
40.5 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 53 | c2hwgA_ |
|
not modelled |
35.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the2 phosphoenolpyruvate:sugar phosphotransferase system
|
| 54 | d1iuka_ |
|
not modelled |
35.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 55 | c3kbqA_ |
|
not modelled |
34.6 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ta0487;
PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum
|
| 56 | c2hroA_ |
|
not modelled |
28.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
| 57 | d2vo1a1 |
|
not modelled |
25.8 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 58 | d3c10a1 |
|
not modelled |
25.8 |
18 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Histone deacetylase, HDAC |
| 59 | c2ohoA_ |
|
not modelled |
25.5 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: structural basis for glutamate racemase inhibitor
|
| 60 | c3rfqC_ |
|
not modelled |
24.8 |
11 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
| 61 | d1xpja_ |
|
not modelled |
24.6 |
11 |
Fold:HAD-like
Superfamily:HAD-like
Family:Hypothetical protein VC0232 |
| 62 | d1s1ma2 |
|
not modelled |
24.3 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 63 | c3fxtB_ |
|
not modelled |
23.7 |
16 |
PDB header:gene regulation
Chain: B: PDB Molecule:nucleoside diphosphate-linked moiety x motif 6;
PDBTitle: crystal structure of the n-terminal domain of human nudt6
|
| 64 | c2ad5B_ |
|
not modelled |
22.3 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: mechanisms of feedback regulation and drug resistance of ctp2 synthetases: structure of the e. coli ctps/ctp complex at 2.8-3 angstrom resolution.
|
| 65 | c1hyhA_ |
|
not modelled |
20.2 |
16 |
PDB header:oxidoreductase (choh(d)-nad+(a))
Chain: A: PDB Molecule:l-2-hydroxyisocaproate dehydrogenase;
PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
|
| 66 | c2ps3A_ |
|
not modelled |
19.1 |
15 |
PDB header:metal transport
Chain: A: PDB Molecule:high-affinity zinc uptake system protein znua;
PDBTitle: structure and metal binding properties of znua, a2 periplasmic zinc transporter from escherichia coli
|
| 67 | c2olsA_ |
|
not modelled |
17.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
|
| 68 | d1hyha1 |
|
not modelled |
15.7 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 69 | d1dxea_ |
|
not modelled |
14.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 70 | d2bdua1 |
|
not modelled |
14.6 |
36 |
Fold:HAD-like
Superfamily:HAD-like
Family:Pyrimidine 5'-nucleotidase (UMPH-1) |
| 71 | d1y81a1 |
|
not modelled |
14.4 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 72 | c3cyvA_ |
|
not modelled |
14.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 shigella flexineri: new insights into its catalytic3 mechanism
|
| 73 | c2bs9B_ |
|
not modelled |
14.0 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-xylosidase;
PDBTitle: native crystal structure of a gh39 beta-xylosidase xynb12 from geobacillus stearothermophilus
|
| 74 | d1c7ga_ |
|
not modelled |
14.0 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 75 | d2d59a1 |
|
not modelled |
13.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 76 | c2csuB_ |
|
not modelled |
13.5 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
| 77 | d1edta_ |
|
not modelled |
13.5 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Type II chitinase |
| 78 | c2vqqA_ |
|
not modelled |
13.4 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:histone deacetylase 4;
PDBTitle: structure of hdac4 catalytic domain (a double cysteine-to-2 alanine mutant) bound to a trifluoromethylketone inhbitor
|
| 79 | d2v1pa1 |
|
not modelled |
12.7 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 80 | c3gycB_ |
|
not modelled |
12.3 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative glycoside hydrolase;
PDBTitle: crystal structure of putative glycoside hydrolase (yp_001304622.1)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
| 81 | d5ldha1 |
|
not modelled |
12.1 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 82 | c2i55C_ |
|
not modelled |
12.0 |
28 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphomannomutase;
PDBTitle: complex of glucose-1,6-bisphosphate with phosphomannomutase from2 leishmania mexicana
|
| 83 | c2v5jB_ |
|
not modelled |
11.5 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;
PDBTitle: apo class ii aldolase hpch
|
| 84 | d1e0ta2 |
|
not modelled |
11.2 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 85 | d1tpla_ |
|
not modelled |
11.2 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 86 | c3i5tB_ |
|
not modelled |
11.2 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase prk07036 from rhodobacter2 sphaeroides kd131
|
| 87 | c2r47C_ |
|
not modelled |
10.4 |
9 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein mth_862;
PDBTitle: crystal structure of mth_862 protein of unknown function from2 methanothermobacter thermautotrophicus
|
| 88 | d2tpsa_ |
|
not modelled |
10.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 89 | d9ldta1 |
|
not modelled |
10.2 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 90 | d1llda1 |
|
not modelled |
10.2 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 91 | c3fcrA_ |
|
not modelled |
10.2 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:putative aminotransferase;
PDBTitle: crystal structure of putative aminotransferase (yp_614685.1) from2 silicibacter sp. tm1040 at 1.80 a resolution
|
| 92 | d1ohwa_ |
|
not modelled |
10.1 |
4 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 93 | c2jfqA_ |
|
not modelled |
9.8 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of staphylococcus aureus glutamate2 racemase in complex with d-glutamate
|
| 94 | c1uhvD_ |
|
not modelled |
9.8 |
10 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-xylosidase;
PDBTitle: crystal structure of beta-d-xylosidase from2 thermoanaerobacterium saccharolyticum, a family 393 glycoside hydrolase
|
| 95 | d1iv0a_ |
|
not modelled |
9.7 |
11 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Putative Holliday junction resolvase RuvX |
| 96 | c2e7vA_ |
|
not modelled |
9.3 |
6 |
PDB header:hydrolase
Chain: A: PDB Molecule:transmembrane protease;
PDBTitle: crystal structure of sea domain of transmembrane protease2 from mus musculus
|
| 97 | c3jszA_ |
|
not modelled |
9.2 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: legionella pneumophila glucosyltransferase lgt1 n293a with udp-glc
|
| 98 | c2wvsD_ |
|
not modelled |
9.0 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|
| 99 | c2vwtA_ |
|
not modelled |
8.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|