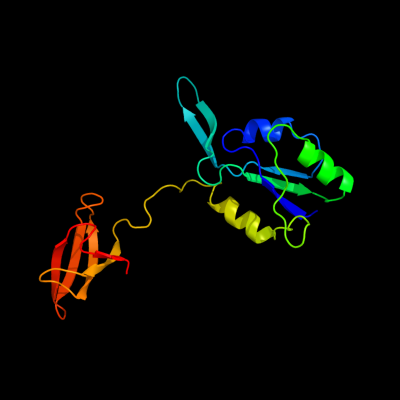
1 c1m1gB_
100.0
21
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of aquifex aeolicus n-utilization2 substance g (nusg), space group p2(1)
2 c2ougC_
100.0
98
PDB header: transcriptionChain: C: PDB Molecule: transcriptional activator rfah;PDBTitle: crystal structure of the rfah transcription factor at 2.1a2 resolution
3 c2xhcA_
100.0
23
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of thermotoga maritima n-utilization substance g2 (nusg)
4 c3p8bB_
99.9
21
PDB header: transferase/transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
5 c2k06A_
99.9
17
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of the aminoterminal domain of e. coli2 nusg
6 d1nz8a_
99.9
26
Fold: Ferredoxin-likeSuperfamily: N-utilization substance G protein NusG, N-terminal domainFamily: N-utilization substance G protein NusG, N-terminal domain7 d1m1ha2
99.9
18
Fold: Ferredoxin-likeSuperfamily: N-utilization substance G protein NusG, N-terminal domainFamily: N-utilization substance G protein NusG, N-terminal domain8 d1nppa2
99.4
19
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain9 d1nz9a_
99.4
21
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain10 c2jvvA_
99.4
20
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
11 c2kvqG_
99.4
20
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
12 c3ewgA_
97.2
17
PDB header: transcriptionChain: A: PDB Molecule: putative transcription antitermination protein nusg;PDBTitle: crystal structure of the n-terminal domain of nusg (ngn) from2 methanocaldococcus jannaschii
13 c2e6zA_
97.0
21
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
14 c2exuA_
96.6
11
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation protein spt4/spt5;PDBTitle: crystal structure of saccharomyces cerevisiae transcription elongation2 factors spt4-spt5ngn domain
15 d2do3a1
96.1
26
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like16 c3h7hB_
95.9
23
PDB header: transcriptionChain: B: PDB Molecule: transcription elongation factor spt5;PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
17 c2ckkA_
95.8
11
PDB header: nuclear proteinChain: A: PDB Molecule: kin17;PDBTitle: high resolution crystal structure of the human kin172 c-terminal domain containing a kow motif3 kin17.
18 c2e70A_
95.6
20
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the fifth kow motif of human2 transcription elongation factor spt5
19 d1vqot1
95.5
20
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e20 c3iz5Y_
95.3
8
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
21 c2zkrt_
not modelled
95.3
10
PDB header: ribosomal protein/rnaChain: T: PDB Molecule: rna expansion segment es39 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
22 c4a1cS_
not modelled
95.1
16
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
23 d2gycs1
not modelled
93.3
16
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e24 d2zjrr1
not modelled
93.2
18
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e25 c2jz2A_
not modelled
90.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ssl0352 protein;PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
26 c1s1iU_
not modelled
85.9
14
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l26-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
27 c3bboW_
not modelled
83.7
17
PDB header: ribosomeChain: W: PDB Molecule: ribosomal protein l24;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
28 c3iz5N_
not modelled
81.4
13
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
29 d2j01y1
not modelled
79.1
21
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e30 c3izcN_
not modelled
79.0
23
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
31 d2joya1
not modelled
77.8
9
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L14e32 c4a19F_
not modelled
69.9
17
PDB header: ribosomeChain: F: PDB Molecule: rpl14;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
33 c2ftcN_
not modelled
67.9
32
PDB header: ribosomeChain: N: PDB Molecule: mitochondrial ribosomal protein l24;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
34 d1ib8a1
not modelled
63.0
10
Fold: Sm-like foldSuperfamily: YhbC-like, C-terminal domainFamily: YhbC-like, C-terminal domain35 d1nxza1
not modelled
51.0
9
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like36 d1vqoq1
not modelled
46.4
19
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e37 c3kbgA_
not modelled
37.8
18
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s4e;PDBTitle: crystal structure of the 30s ribosomal protein s4e from2 thermoplasma acidophilum. northeast structural genomics3 consortium target tar28.
38 c4a18N_
not modelled
36.9
17
PDB header: ribosomeChain: N: PDB Molecule: rpl27;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
39 c3qiiA_
not modelled
36.5
10
PDB header: transcription regulatorChain: A: PDB Molecule: phd finger protein 20;PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
40 d1rkia1
not modelled
31.3
10
Fold: THUMP domainSuperfamily: THUMP domain-likeFamily: PAE0736-like41 d2vv5a1
not modelled
29.8
14
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Mechanosensitive channel protein MscS (YggB), middle domain42 d2pu9b1
not modelled
29.0
18
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Ferredoxin thioredoxin reductase (FTR), alpha (variable) chain43 d1vhka1
not modelled
28.0
14
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like44 d2zgwa1
not modelled
27.9
34
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Biotin repressor (BirA)45 d1h3ga1
not modelled
25.2
36
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes46 c2dxcG_
not modelled
24.0
19
PDB header: hydrolaseChain: G: PDB Molecule: thiocyanate hydrolase subunit alpha;PDBTitle: recombinant thiocyanate hydrolase, fully-matured form
47 c3ikmD_
not modelled
23.0
27
PDB header: transferaseChain: D: PDB Molecule: dna polymerase subunit gamma-1;PDBTitle: crystal structure of human mitochondrial dna polymerase2 holoenzyme
48 c3iz6D_
not modelled
22.5
16
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein s4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
49 c3pifD_
not modelled
21.4
17
PDB header: hydrolaseChain: D: PDB Molecule: 5'->3' exoribonuclease (xrn1);PDBTitle: crystal structure of the 5'->3' exoribonuclease xrn1, e178q mutant in2 complex with manganese
50 c2gpzC_
not modelled
21.1
17
PDB header: hydrolaseChain: C: PDB Molecule: transthyretin-like protein;PDBTitle: transthyretin-like protein from salmonella dublin
51 c3j04A_
not modelled
17.9
13
PDB header: structural proteinChain: A: PDB Molecule: myosin-11;PDBTitle: em structure of the heavy meromyosin subfragment of chick smooth2 muscle myosin with regulatory light chain in phosphorylated state
52 d1a8pa1
not modelled
17.7
13
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like53 c3d8tB_
not modelled
17.5
17
PDB header: lyaseChain: B: PDB Molecule: uroporphyrinogen-iii synthase;PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
54 c2h1xB_
not modelled
17.3
11
PDB header: hydrolaseChain: B: PDB Molecule: 5-hydroxyisourate hydrolase (formerly known asPDBTitle: crystal structure of 5-hydroxyisourate hydrolase (formerly2 known as trp, transthyretin related protein)
55 c2vc8A_
not modelled
16.6
20
PDB header: protein-bindingChain: A: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of the lsm domain of human edc3 (enhancer2 of decapping 3)
56 d1yt8a4
not modelled
16.6
12
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)57 d1ugpb_
not modelled
15.5
24
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain58 d1wd7a_
not modelled
15.4
17
Fold: HemD-likeSuperfamily: HemD-likeFamily: HemD-like59 c2k3aA_
not modelled
15.2
18
PDB header: hydrolaseChain: A: PDB Molecule: chap domain protein;PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
60 c2equA_
not modelled
14.9
10
PDB header: protein bindingChain: A: PDB Molecule: phd finger protein 20-like 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
61 d1v29b_
not modelled
14.7
10
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain62 c2xzmW_
not modelled
14.2
14
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s4;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
63 d2hqha1
not modelled
14.0
14
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain64 c3izcG_
not modelled
13.9
18
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein rpl6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
65 c2vv5D_
not modelled
13.6
14
PDB header: membrane proteinChain: D: PDB Molecule: small-conductance mechanosensitive channel;PDBTitle: the open structure of mscs
66 c3p8dB_
not modelled
13.4
7
PDB header: protein bindingChain: B: PDB Molecule: medulloblastoma antigen mu-mb-50.72;PDBTitle: crystal structure of the second tudor domain of human phf20 (homodimer2 form)
67 d1jr2a_
not modelled
12.3
24
Fold: HemD-likeSuperfamily: HemD-likeFamily: HemD-like68 c1jr2A_
not modelled
12.3
24
PDB header: lyaseChain: A: PDB Molecule: uroporphyrinogen-iii synthase;PDBTitle: structure of uroporphyrinogen iii synthase
69 d1ixda_
not modelled
12.3
20
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain70 c1x60A_
not modelled
12.1
3
PDB header: hydrolaseChain: A: PDB Molecule: sporulation-specific n-acetylmuramoyl-l-alaninePDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
71 d1f86a_
not modelled
12.1
0
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)72 d2qdyb1
not modelled
12.0
15
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain73 d1s7ia_
not modelled
11.9
29
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: DGPF domain (Pfam 04946)74 d1kgia_
not modelled
11.5
0
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)75 d2coya1
not modelled
11.3
11
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain76 c2hqxB_
not modelled
10.7
11
PDB header: transcriptionChain: B: PDB Molecule: p100 co-activator tudor domain;PDBTitle: crystal structure of human p100 tudor domain conserved2 region
77 d2hqxa1
not modelled
10.7
11
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain78 c2rm4A_
not modelled
10.6
20
PDB header: protein bindingChain: A: PDB Molecule: cg6311-pb;PDBTitle: solution structure of the lsm domain of dm edc3 (enhancer2 of decapping 3)
79 d1e0ta3
not modelled
10.5
12
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain80 d2diqa1
not modelled
10.3
11
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain81 c2ky9A_
not modelled
10.3
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ydhk;PDBTitle: solution nmr structure of ydhk c-terminal domain from b.subtilis,2 northeast structural genomics consortium target target sr518
82 c3qz9D_
not modelled
10.1
20
PDB header: lyaseChain: D: PDB Molecule: co-type nitrile hydratase beta subunit;PDBTitle: crystal structure of co-type nitrile hydratase beta-y215f from2 pseudomonas putida.
83 d2p3ha1
not modelled
10.1
10
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like84 d2hlja1
not modelled
10.0
11
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like85 c1nwxN_
not modelled
9.5
20
PDB header: ribosomeChain: N: PDB Molecule: ribosomal protein l19;PDBTitle: complex of the large ribosomal subunit from deinococcus2 radiodurans with abt-773
86 c3izca_
not modelled
9.4
18
PDB header: ribosomeChain: A: PDB Molecule: 60s ribosomal protein rpl1 (l1p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
87 d1fdra1
not modelled
9.4
33
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like88 d1ttaa_
not modelled
9.3
0
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)89 d2zjrm1
not modelled
9.2
13
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L1990 c3izbD_
not modelled
9.1
16
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein rps4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
91 c1vhkA_
not modelled
9.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yqeu;PDBTitle: crystal structure of an hypothetical protein
92 c2eqmA_
not modelled
9.0
11
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 20-like 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1 [homo sapiens]
93 d1rw1a_
not modelled
8.9
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: ArsC-like94 c1i84V_
not modelled
8.9
14
PDB header: contractile proteinChain: V: PDB Molecule: smooth muscle myosin heavy chain;PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.
95 d1oo2a_
not modelled
8.8
6
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)96 c3nnhA_
not modelled
8.5
17
PDB header: rna binding protein/rnaChain: A: PDB Molecule: cugbp elav-like family member 1;PDBTitle: crystal structure of the cugbp1 rrm1 with guuguuuuguuu rna
97 c3q1jA_
not modelled
8.4
10
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 20;PDBTitle: crystal structure of tudor domain 1 of human phd finger protein 20
98 c2k3yA_
not modelled
8.4
14
PDB header: transcription regulatorChain: A: PDB Molecule: chromatin modification-related protein eaf3;PDBTitle: solution structure of eaf3 chromo barrel domain bound to2 histone h3 with a dimethyllysine analog h3k36me2
99 c2pmzV_
not modelled
8.3
13
PDB header: translation, transferaseChain: V: PDB Molecule: dna-directed rna polymerase subunit h;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus