1 c3dh3C_
100.0
100
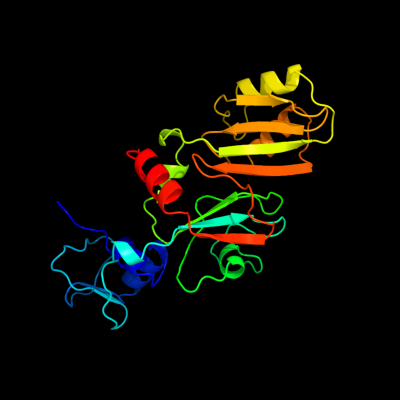
PDB header: isomerase/rnaChain: C: PDB Molecule: ribosomal large subunit pseudouridine synthase f;PDBTitle: crystal structure of rluf in complex with a 22 nucleotide2 rna substrate
2 c1kskA_
100.0
28
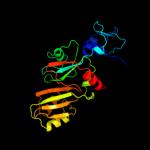
PDB header: lyaseChain: A: PDB Molecule: ribosomal small subunit pseudouridine synthase a;PDBTitle: structure of rsua
3 c1vioA_
100.0
31
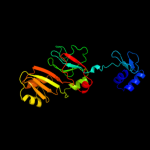
PDB header: lyaseChain: A: PDB Molecule: ribosomal small subunit pseudouridine synthase a;PDBTitle: crystal structure of pseudouridylate synthase
4 c2gmlA_
100.0
98
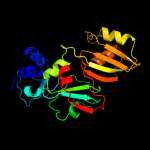
PDB header: isomeraseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase f;PDBTitle: crystal structure of catalytic domain of e.coli rluf
5 d1kska4
100.0
29
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD6 d1vioa1
100.0
31
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD7 c2omlA_
100.0
35
PDB header: isomeraseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase e;PDBTitle: crystal structure of e. coli pseudouridine synthase rlue
8 c2olwB_
100.0
33
PDB header: isomeraseChain: B: PDB Molecule: ribosomal large subunit pseudouridine synthase e;PDBTitle: crystal structure of e. coli pseudouridine synthase rlue
9 d1v9ka_
100.0
19
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD10 c2i82D_
99.9
15
PDB header: lyase/rnaChain: D: PDB Molecule: ribosomal large subunit pseudouridine synthase a;PDBTitle: crystal structure of pseudouridine synthase rlua: indirect2 sequence readout through protein-induced rna structure
11 c1v9fA_
99.9
20
PDB header: lyaseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase d;PDBTitle: crystal structure of catalytic domain of pseudouridine2 synthase rlud from escherichia coli
12 d1v9fa_
99.9
20
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD13 c1qyuA_
99.9
20
PDB header: lyaseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase d;PDBTitle: structure of the catalytic domain of 23s rrna pseudouridine2 synthase rlud
14 d1vioa2
99.4
29
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Pseudouridine synthase RsuA N-terminal domain15 d1p9ka_
99.3
21
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: YbcJ-like16 d1dm9a_
99.2
29
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Heat shock protein 15 kD17 c1dm9A_
99.2
29
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical 15.5 kd protein in mrca-pckaPDBTitle: heat shock protein 15 kd
18 c3bbnD_
99.0
22
PDB header: ribosomeChain: D: PDB Molecule: ribosomal protein s4;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
19 c2k6pA_
99.0
27
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein hp_1423;PDBTitle: solution structure of hypothetical protein, hp1423
20 d1c06a_
99.0
24
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Ribosomal protein S421 c2cqjA_
not modelled
99.0
22
PDB header: rna binding proteinChain: A: PDB Molecule: u3 small nucleolar ribonucleoprotein proteinPDBTitle: solution structure of the s4 domain of u3 small nucleolar2 ribonucleoprotein protein imp3 homolog
22 d2uubd1
not modelled
98.9
31
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Ribosomal protein S423 d2gy9d1
not modelled
98.7
27
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Ribosomal protein S424 d2apoa2
not modelled
98.6
22
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB25 d2ey4a2
not modelled
98.6
21
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB26 c2ey4A_
not modelled
98.5
23
PDB header: isomerase/biosynthetic proteinChain: A: PDB Molecule: probable trna pseudouridine synthase b;PDBTitle: crystal structure of a cbf5-nop10-gar1 complex
27 d1r3ea2
not modelled
98.5
27
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB28 d1k8wa5
not modelled
98.5
20
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB29 c2apoA_
not modelled
98.4
21
PDB header: isomerase/rna binding proteinChain: A: PDB Molecule: probable trna pseudouridine synthase b;PDBTitle: crystal structure of the methanococcus jannaschii cbf52 nop10 complex
30 d1sgva2
not modelled
98.3
22
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB31 c1sgvA_
not modelled
98.2
22
PDB header: lyaseChain: A: PDB Molecule: trna pseudouridine synthase b;PDBTitle: structure of trna psi55 pseudouridine synthase (trub)
32 c3uaiA_
not modelled
98.2
19
PDB header: isomerase/chaperoneChain: A: PDB Molecule: h/aca ribonucleoprotein complex subunit 4;PDBTitle: structure of the shq1-cbf5-nop10-gar1 complex from saccharomyces2 cerevisiae
33 c1k8wA_
not modelled
98.1
18
PDB header: lyase/rnaChain: A: PDB Molecule: trna pseudouridine synthase b;PDBTitle: crystal structure of the e. coli pseudouridine synthase2 trub bound to a t stem-loop rna
34 c3hp7A_
not modelled
98.1
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hemolysin, putative;PDBTitle: putative hemolysin from streptococcus thermophilus.
35 d1kska3
not modelled
97.9
26
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Pseudouridine synthase RsuA N-terminal domain36 c1s1hD_
not modelled
97.6
16
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein s9-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
37 c2xzmD_
not modelled
97.6
12
PDB header: ribosomeChain: D: PDB Molecule: ribosomal protein s4 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
38 d1jh3a_
not modelled
97.5
22
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Tyrosyl-tRNA synthetase (TyrRS), C-terminal domain39 d1h3fa2
not modelled
97.5
23
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Tyrosyl-tRNA synthetase (TyrRS), C-terminal domain40 c2janD_
not modelled
97.1
22
PDB header: ligaseChain: D: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: tyrosyl-trna synthetase from mycobacterium tuberculosis in2 unliganded state
41 c1h3eA_
not modelled
97.0
23
PDB header: ligaseChain: A: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: tyrosyl-trna synthetase from thermus thermophilus complexed2 with wild-type trnatyr(gua) and with atp and tyrosinol
42 c1ze2B_
not modelled
93.9
23
PDB header: lyase/rnaChain: B: PDB Molecule: trna pseudouridine synthase b;PDBTitle: conformational change of pseudouridine 55 synthase upon its2 association with rna substrate
43 c3iz6C_
not modelled
93.6
17
PDB header: ribosomeChain: C: PDB Molecule: 40s ribosomal protein s9 (s4p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
44 c3kbgA_
not modelled
93.5
26
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s4e;PDBTitle: crystal structure of the 30s ribosomal protein s4e from2 thermoplasma acidophilum. northeast structural genomics3 consortium target tar28.
45 c2xzmW_
not modelled
92.8
23
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s4;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
46 c3iz6D_
not modelled
91.2
23
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein s4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
47 c3izbD_
not modelled
91.0
24
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein rps4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
48 d2g1la1
not modelled
70.3
24
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain49 d1fm0d_
not modelled
64.5
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD50 c3fm8A_
not modelled
63.3
24
PDB header: transport protein/hydrolase activatorChain: A: PDB Molecule: kinesin-like protein kif13b;PDBTitle: crystal structure of full length centaurin alpha-1 bound with the fha2 domain of kif13b (capri target)
51 c3hvzB_
not modelled
59.6
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
52 c2eh0A_
not modelled
58.3
24
PDB header: transport proteinChain: A: PDB Molecule: kinesin-like protein kif1b;PDBTitle: solution structure of the fha domain from human kinesin-2 like protein kif1b
53 c3h7hA_
not modelled
58.3
22
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt4;PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
54 c3po0A_
not modelled
58.1
35
PDB header: protein bindingChain: A: PDB Molecule: small archaeal modifier protein 1;PDBTitle: crystal structure of samp1 from haloferax volcanii
55 c2qieB_
not modelled
55.9
26
PDB header: transferaseChain: B: PDB Molecule: molybdopterin synthase small subunit;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
56 d1rwsa_
not modelled
55.8
29
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS57 c2kmmA_
not modelled
54.9
29
PDB header: hydrolaseChain: A: PDB Molecule: guanosine-3',5'-bis(diphosphate) 3'-PDBTitle: solution nmr structure of the tgs domain of pg1808 from2 porphyromonas gingivalis. northeast structural genomics3 consortium target pgr122a (418-481)
58 d1tkea1
not modelled
48.7
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain59 d2hzab1
not modelled
47.9
22
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like60 d1wlna1
not modelled
47.3
12
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain61 d1vjka_
not modelled
46.5
39
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD62 d2hzaa1
not modelled
44.9
23
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like63 c1z4hA_
not modelled
43.9
27
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: tor inhibition protein;PDBTitle: the response regulator tori belongs to a new family of2 atypical excisionase
64 d2affa1
not modelled
42.2
24
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain65 c3gmgB_
not modelled
41.3
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rv1825/mt1873;PDBTitle: crystal structure of an uncharacterized conserved protein2 from mycobacterium tuberculosis
66 c2exuA_
not modelled
39.0
17
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation protein spt4/spt5;PDBTitle: crystal structure of saccharomyces cerevisiae transcription elongation2 factors spt4-spt5ngn domain
67 d2ff4a3
not modelled
35.4
11
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain68 c3dwmA_
not modelled
34.4
18
PDB header: transferaseChain: A: PDB Molecule: 9.5 kda culture filtrate antigen cfp10a;PDBTitle: crystal structure of mycobacterium tuberculosis cyso, an antigen
69 d1nyra2
not modelled
33.8
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain70 c1q5vB_
not modelled
33.4
24
PDB header: transcriptionChain: B: PDB Molecule: nickel responsive regulator;PDBTitle: apo-nikr
71 c2bj3D_
not modelled
32.6
28
PDB header: transcriptionChain: D: PDB Molecule: nickel responsive regulator;PDBTitle: nikr-apo
72 c1r21A_
not modelled
31.6
17
PDB header: cell cycleChain: A: PDB Molecule: antigen ki-67;PDBTitle: solution structure of human ki67 fha domain
73 c3rpfC_
not modelled
30.8
21
PDB header: transferaseChain: C: PDB Molecule: molybdopterin converting factor, subunit 1 (moad);PDBTitle: protein-protein complex of subunit 1 and 2 of molybdopterin-converting2 factor from helicobacter pylori 26695
74 d1wxqa2
not modelled
30.1
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain75 d1xo3a_
not modelled
27.5
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog76 c2qjlA_
not modelled
27.1
20
PDB header: signaling proteinChain: A: PDB Molecule: ubiquitin-related modifier 1;PDBTitle: crystal structure of urm1
77 c3gqsB_
not modelled
26.7
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: adenylate cyclase-like protein;PDBTitle: crystal structure of the fha domain of ct664 protein from chlamydia2 trachomatis
78 d1zud21
not modelled
25.1
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS79 d2cu3a1
not modelled
24.8
36
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS80 c2g1eA_
not modelled
24.6
22
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein ta0895;PDBTitle: solution structure of ta0895
81 d1v8ca1
not modelled
24.4
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD82 c1wwtA_
not modelled
24.3
5
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase, cytoplasmic;PDBTitle: solution structure of the tgs domain from human threonyl-2 trna synthetase
83 d1c1za5
not modelled
24.0
21
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain84 c3poaA_
not modelled
23.9
20
PDB header: peptide binding proteinChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: structural and functional analysis of phosphothreonine-dependent fha2 domain interactions
85 d2piea1
not modelled
23.4
21
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain86 d2bj7a1
not modelled
23.1
38
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like87 c2jqlA_
not modelled
22.2
28
PDB header: cell cycleChain: A: PDB Molecule: dna damage response protein kinase dun1;PDBTitle: nmr structure of the yeast dun1 fha domain in complex with2 a doubly phosphorylated (pt) peptide derived from rad533 scd1
88 c3kt9A_
not modelled
22.1
14
PDB header: hydrolaseChain: A: PDB Molecule: aprataxin;PDBTitle: aprataxin fha domain
89 c2kl0A_
not modelled
21.4
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
90 c2ca9B_
not modelled
21.3
28
PDB header: transcriptional regulationChain: B: PDB Molecule: putative nickel-responsive regulator;PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
91 c1yj5C_
not modelled
21.0
16
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
92 c3cwiA_
not modelled
20.7
41
PDB header: biosynthetic proteinChain: A: PDB Molecule: thiamine-biosynthesis protein this;PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
93 c2r9qD_
not modelled
20.5
9
PDB header: hydrolaseChain: D: PDB Molecule: 2'-deoxycytidine 5'-triphosphate deaminase;PDBTitle: crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from2 agrobacterium tumefaciens
94 c1tygG_
not modelled
20.2
9
PDB header: biosynthetic proteinChain: G: PDB Molecule: yjbs;PDBTitle: structure of the thiazole synthase/this complex
95 c1f5nA_
not modelled
19.5
13
PDB header: signaling proteinChain: A: PDB Molecule: interferon-induced guanylate-binding protein 1;PDBTitle: human guanylate binding protein-1 in complex with the gtp2 analogue, gmppnp.
96 d1ryja_
not modelled
18.5
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS97 d1xnea_
not modelled
18.2
19
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain98 c2l52A_
not modelled
18.1
22
PDB header: protein bindingChain: A: PDB Molecule: methanosarcina acetivorans samp1 homolog;PDBTitle: solution structure of the small archaeal modifier protein 1 (samp1)2 from methanosarcina acetivorans
99 d1wgka_
not modelled
18.0
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog