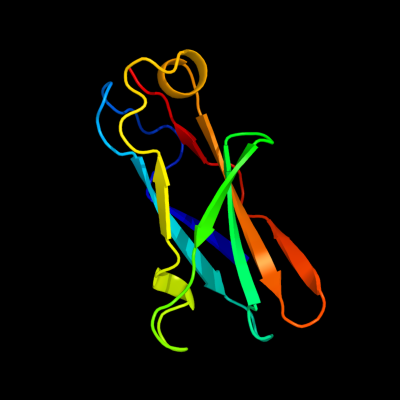
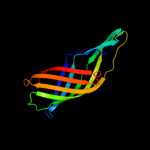
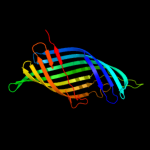
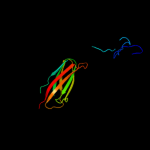
1 c3pdgA_
97.0
13
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
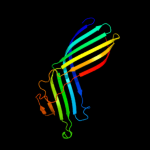
2 c3pe9A_
97.0
8
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
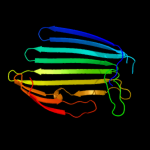
3 c3pe9C_
96.9
8
PDB header: unknown functionChain: C: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
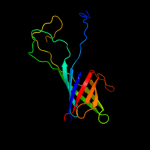
4 c3pe9D_
96.5
9
PDB header: unknown functionChain: D: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
5 c3pe9B_
96.1
8
PDB header: unknown functionChain: B: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
6 c3nb3C_
95.9
19
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
7 d1qj8a_
95.7
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein8 c3qraA_
94.6
15
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
9 d1phoa_
94.2
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin10 c3pddA_
93.0
11
PDB header: unknown functionChain: A: PDB Molecule: glycoside hydrolase, family 9;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
11 d1osma_
92.9
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin12 c2f1tB_
92.6
15
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
13 c2x4mD_
92.5
17
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
14 d2zfga1
91.9
12
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin15 c2k0lA_
91.6
19
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
16 c2jmmA_
90.9
14
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
17 d1t16a_
90.1
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Outer membrane protein transport protein18 d1g90a_
89.8
18
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein19 d1qjpa_
89.8
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein20 c2qomB_
89.7
13
PDB header: hydrolaseChain: B: PDB Molecule: serine protease espp;PDBTitle: the crystal structure of the e.coli espp autotransporter beta-domain.
21 c3bryB_
not modelled
88.4
9
PDB header: transport proteinChain: B: PDB Molecule: tbux;PDBTitle: crystal structure of the ralstonia pickettii toluene2 transporter tbux
22 c3brzA_
not modelled
87.7
11
PDB header: transport proteinChain: A: PDB Molecule: todx;PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
23 c3aehB_
not modelled
87.3
10
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp autotransporter;PDBTitle: integral membrane domain of autotransporter hbp
24 d1uynx_
not modelled
81.7
11
Fold: Transmembrane beta-barrelsSuperfamily: AutotransporterFamily: Autotransporter25 c2yrlA_
not modelled
81.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kiaa1837 protein;PDBTitle: solution structure of the pkd domain from kiaa 1837 protein
26 c2x27X_
not modelled
80.7
12
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
27 d1p4ta_
not modelled
79.2
13
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein28 d1i78a_
not modelled
77.2
12
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane protease OMPT29 d1wgoa_
not modelled
75.7
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain30 d2fcba2
not modelled
72.2
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains31 d1l0qa1
not modelled
71.8
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain32 c3dwoX_
not modelled
71.5
14
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
33 c2kzwA_
not modelled
68.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of q8psa4 from methanosarcina mazei, northeast2 structural genomics consortium target mar143a
34 d1fnla2
not modelled
67.9
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains35 d1f2qa2
not modelled
65.4
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains36 c3nsgA_
not modelled
61.5
13
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
37 c3qq2C_
not modelled
61.4
12
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
38 c2lhfA_
not modelled
59.1
16
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein h1;PDBTitle: solution structure of outer membrane protein h (oprh) from p.2 aeruginosa in dhpc micelles
39 c2e7mA_
not modelled
58.0
12
PDB header: structural proteinChain: A: PDB Molecule: protein kiaa0319;PDBTitle: solution structure of the pkd domain (329-428) from human2 kiaa0319
40 c3o3uN_
not modelled
57.5
15
PDB header: transport protein, signaling proteinChain: N: PDB Molecule: maltose-binding periplasmic protein, advanced glycosylationPDBTitle: crystal structure of human receptor for advanced glycation endproducts2 (rage)
41 d1epfa1
not modelled
56.2
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains42 c2a74B_
not modelled
55.8
9
PDB header: immune systemChain: B: PDB Molecule: complement component c3c;PDBTitle: human complement component c3c
43 d1pama3
not modelled
55.6
19
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain44 d1w8oa1
not modelled
55.5
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes45 c2xwxB_
53.3
17
PDB header: chitin-binding proteinChain: B: PDB Molecule: glcnac-binding protein a;PDBTitle: vibrio cholerae colonization factor gbpa crystal structure
46 c2e7bA_
not modelled
51.4
13
PDB header: structural proteinChain: A: PDB Molecule: obscurin;PDBTitle: solution structure of the 6th ig-like domain from human2 kiaa1556
47 c2c26A_
not modelled
50.6
21
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: structural basis for the promiscuous specificity of the2 carbohydrate-binding modules from the beta-sandwich super3 family
48 c2ensA_
not modelled
50.5
22
PDB header: signaling proteinChain: A: PDB Molecule: advanced glycosylation end product-specificPDBTitle: solution structure of the third ig-like domain from human2 advanced glycosylation end product-specific receptor
49 d3bmva3
not modelled
50.5
24
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain50 d1qhoa3
not modelled
49.7
19
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain51 c3ubfA_
not modelled
49.1
19
PDB header: cell adhesionChain: A: PDB Molecule: neural-cadherin;PDBTitle: crystal structure of drosophila n-cadherin ec1-3, i
52 c3jquA_
not modelled
48.5
14
PDB header: cell adhesionChain: A: PDB Molecule: collagenase;PDBTitle: crystal structure of clostridium histolyticum colg collagenase2 polycystic kidney disease domain at 1.4 angstrom resolution
53 d1cyga3
not modelled
47.2
19
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain54 d1cxla3
not modelled
46.4
22
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain55 d1biha3
not modelled
46.1
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains56 d1tiua_
not modelled
45.2
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains57 c2ee0A_
not modelled
45.1
9
PDB header: signaling proteinChain: A: PDB Molecule: protocadherin-9;PDBTitle: solution structures of the ca domain of human protocadherin2 9
58 d2fdbp2
not modelled
44.6
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains59 d1cgta3
not modelled
43.8
15
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain60 c3ottB_
not modelled
43.0
14
PDB header: transcriptionChain: B: PDB Molecule: two-component system sensor histidine kinase;PDBTitle: crystal structure of the extracellular domain of the putative one2 component system bt4673 from b. thetaiotaomicron
61 c2wjqA_
not modelled
41.9
11
PDB header: transport proteinChain: A: PDB Molecule: probable n-acetylneuraminic acid outer membrane channelPDBTitle: nanc porin structure in hexagonal crystal form.
62 d1fcga2
not modelled
41.9
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains63 c2k1mA_
not modelled
41.4
11
PDB header: structural proteinChain: A: PDB Molecule: myosin-binding protein c, cardiac-type;PDBTitle: 3d nmr structure of domain cc0 of cardiac myosin binding2 protein c (mybpc)
64 c2o4vA_
not modelled
40.9
14
PDB header: membrane proteinChain: A: PDB Molecule: porin p;PDBTitle: an arginine ladder in oprp mediates phosphate specific transfer across2 the outer membrane
65 c3ff7B_
not modelled
40.6
16
PDB header: cell adhesion/immunue systemChain: B: PDB Molecule: epithelial cadherin;PDBTitle: structure of nk cell receptor klrg1 bound to e-cadherin
66 c1l0qC_
not modelled
40.0
10
PDB header: protein bindingChain: C: PDB Molecule: surface layer protein;PDBTitle: tandem yvtn beta-propeller and pkd domains from an archaeal surface2 layer protein
67 c3s37X_
not modelled
39.8
12
PDB header: immune system/transferaseChain: X: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: structural basis for the function of two anti-vegf receptor antibodies
68 c2enyA_
not modelled
39.6
11
PDB header: contractile proteinChain: A: PDB Molecule: obscurin;PDBTitle: solution structure of the ig-like domain (2735-2825) of2 human obscurin
69 c3ppeB_
not modelled
39.6
16
PDB header: cell adhesionChain: B: PDB Molecule: vascular endothelial cadherin;PDBTitle: crystal structure of chicken ve-cadherin ec1-2
70 c2cr6A_
not modelled
37.9
13
PDB header: contractile proteinChain: A: PDB Molecule: kiaa1556 protein;PDBTitle: solution structure of the ig domain (2998-3100) of human2 obscurin
71 c2gqhA_
not modelled
37.4
12
PDB header: contractile proteinChain: A: PDB Molecule: kiaa1556 protein;PDBTitle: solution structure of the 15th ig-like domain of human2 kiaa1556 protein
72 c2jjsB_
not modelled
37.4
10
PDB header: cell adhesionChain: B: PDB Molecule: tyrosine-protein phosphatase non-receptor typePDBTitle: structure of human cd47 in complex with human signal2 regulatory protein (sirp) alpha
73 d2fgqx1
not modelled
36.9
15
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin74 d1epfa2
not modelled
36.8
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains75 c2cknA_
not modelled
36.6
13
PDB header: transferaseChain: A: PDB Molecule: basic fibroblast growth factor receptor 1;PDBTitle: nmr structure of the first ig module of mouse fgfr1
76 c2dkuA_
not modelled
36.6
11
PDB header: contractile proteinChain: A: PDB Molecule: kiaa1556 protein;PDBTitle: solution structure of the third ig-like domain of human2 kiaa1556 protein
77 d1gl4b_
not modelled
36.1
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains78 d2nqda1
not modelled
35.2
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ICP-likeFamily: ICP-like79 c2nmsA_
not modelled
34.3
9
PDB header: immune systemChain: A: PDB Molecule: cmrf35-like-molecule 1;PDBTitle: the crystal structure of the extracellular domain of the2 inhibitor receptor expressed on myeloid cells irem-1
80 d1l3wa5
not modelled
33.5
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Cadherin81 c2e6pA_
not modelled
32.9
14
PDB header: structural proteinChain: A: PDB Molecule: obscurin-like protein 1;PDBTitle: solution structure of the ig-like domain (714-804) from2 human obscurin-like protein 1
82 d1pd6a_
not modelled
31.9
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains83 d1l3wa4
not modelled
31.9
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Cadherin84 c3hs0B_
not modelled
31.8
11
PDB header: immune systemChain: B: PDB Molecule: cobra venom factor;PDBTitle: cobra venom factor (cvf) in complex with human factor b
85 c2bk8A_
not modelled
31.2
14
PDB header: ig domainChain: A: PDB Molecule: titin heart isoform n2-b;PDBTitle: m1 domain from titin
86 c1ncjA_
not modelled
30.4
13
PDB header: cell adhesion proteinChain: A: PDB Molecule: protein (n-cadherin);PDBTitle: n-cadherin, two-domain fragment
87 c1gzpA_
not modelled
30.1
15
PDB header: glycoproteinChain: A: PDB Molecule: t-cell surface glycoprotein cd1b;PDBTitle: cd1b in complex with gm2 ganglioside
88 d1o75a1
not modelled
29.4
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Tp47 lipoprotein, middle and C-terminal domainsFamily: Tp47 lipoprotein, middle and C-terminal domains89 c1nchB_
not modelled
28.8
16
PDB header: cell adhesion proteinChain: B: PDB Molecule: n-cadherin;PDBTitle: structural basis of cell-cell adhesion by cadherins
90 d1f97a2
not modelled
28.8
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains91 c3m86B_
not modelled
28.6
17
PDB header: protein bindingChain: B: PDB Molecule: amoebiasin-2;PDBTitle: crystal structure of the cysteine protease inhibitor, ehicp2, from2 entamoeba histolytica
92 c1bqsA_
not modelled
27.6
16
PDB header: membrane proteinChain: A: PDB Molecule: protein (mucosal addressin cell adhesionPDBTitle: the crystal structure of mucosal addressin cell adhesion2 molecule-1 (madcam-1)
93 c3ff8B_
not modelled
27.6
14
PDB header: cell adhesion/immune systemChain: B: PDB Molecule: epithelial cadherin;PDBTitle: structure of nk cell receptor klrg1 bound to e-cadherin
94 c1q5cA_
not modelled
26.6
10
PDB header: structural proteinChain: A: PDB Molecule: ep-cadherin;PDBTitle: s-s-lambda-shaped trans and cis interactions of cadherins model based2 on fitting c-cadherin (1l3w) to 3d map of desmosomes obtained by3 electron tomography
95 c2edoA_
not modelled
26.0
14
PDB header: cell adhesionChain: A: PDB Molecule: cd48 antigen;PDBTitle: solution structure of the first ig-like domain from human2 cd48 antigen
96 c3a2rX_
not modelled
25.9
15
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein ii;PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
97 d1tlya_
not modelled
25.7
19
Fold: Transmembrane beta-barrelsSuperfamily: Tsx-like channelFamily: Tsx-like channel98 c2y25C_
not modelled
25.7
14
PDB header: structural proteinChain: C: PDB Molecule: myomesin;PDBTitle: crystal structure of the myomesin domains my11-my13
99 c2oxgE_
not modelled
25.1
19
PDB header: transport proteinChain: E: PDB Molecule: soxz protein;PDBTitle: the soxyz complex of paracoccus pantotrophus
100 c2yuvA_
not modelled
25.0
11
PDB header: cell adhesionChain: A: PDB Molecule: myosin-binding protein c, slow-type;PDBTitle: solution structure of 2nd immunoglobulin domain of slow2 type myosin-binding protein c
101 d1l3wa1
not modelled
24.9
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Cadherin102 c2nplX_
not modelled
24.7
13
PDB header: cell adhesionChain: X: PDB Molecule: coxsackievirus and adenovirus receptor;PDBTitle: nmr structure of card d2 domain
103 d1b4ra_
not modelled
24.5
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain104 d1biha1
not modelled
23.1
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains105 c3g6jB_
not modelled
23.0
9
PDB header: immune systemChain: B: PDB Molecule: complement c3 alpha chain;PDBTitle: c3b in complex with a c3b specific fab
106 c2if7C_
not modelled
22.8
21
PDB header: immune systemChain: C: PDB Molecule: slam family member 6;PDBTitle: crystal structure of ntb-a
107 d1u2ca1
not modelled
22.6
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Dystroglycan, N-terminal domain108 c1zvnA_
not modelled
22.2
13
PDB header: ----Chain: A: PDB Molecule: mn-cadherin;PDBTitle: crystal structure of chick mn-cadherin ec1
109 c2yuzA_
not modelled
21.7
7
PDB header: immune systemChain: A: PDB Molecule: myosin-binding protein c, slow-type;PDBTitle: solution structure of 4th immunoglobulin domain of slow2 type myosin-binding protein c
110 c1hngB_
not modelled
21.5
15
PDB header: t lymphocyte adhesion glycoproteinChain: B: PDB Molecule: cd2;PDBTitle: crystal structure at 2.8 angstroms resolution of a soluble2 form of the cell adhesion molecule cd2
111 c2a73B_
not modelled
21.3
10
PDB header: immune systemChain: B: PDB Molecule: complement c3;PDBTitle: human complement component c3
112 c3lndC_
not modelled
21.2
19
PDB header: cell adhesionChain: C: PDB Molecule: cdh6 protein;PDBTitle: crystal structure of cadherin-6 ec12 w4a
113 c3mpcB_
not modelled
20.9
19
PDB header: unknown functionChain: B: PDB Molecule: fn3-like protein;PDBTitle: the crystal structure of a fn3-like protein from clostridium2 thermocellum
114 c1nciA_
not modelled
20.8
16
PDB header: cell adhesion proteinChain: A: PDB Molecule: n-cadherin;PDBTitle: structural basis of cell-cell adhesion by cadherins
115 d1ncia_
not modelled
20.8
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Cadherin116 c2wd0C_
not modelled
20.2
9
PDB header: cell adhesionChain: C: PDB Molecule: cadherin-23;PDBTitle: crystal structure of nonsyndromic deafness (dfnb12)2 associated mutant d124g of mouse cadherin-23 ec1-2