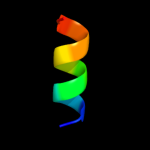
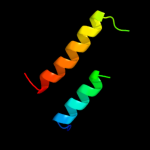
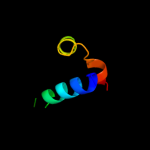
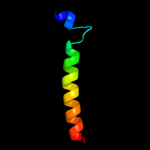
1 c1ciiA_
66.2
14
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
2 c3fewX_
59.7
17
PDB header: immune systemChain: X: PDB Molecule: colicin s4;PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
3 d1yqga1
51.0
12
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: ProC C-terminal domain-like4 d1cola_
45.0
17
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin5 d1rh1a2
44.9
10
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin6 c1rh1A_
28.1
10
PDB header: antibioticChain: A: PDB Molecule: colicin b;PDBTitle: crystal structure of the cytotoxic bacterial protein2 colicin b at 2.5 a resolution
7 d1uxca_
27.5
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator8 c2ag8A_
21.6
12
PDB header: oxidoreductaseChain: A: PDB Molecule: pyrroline-5-carboxylate reductase;PDBTitle: nadp complex of pyrroline-5-carboxylate reductase from neisseria2 meningitidis
9 c3mb2J_
20.4
26
PDB header: isomeraseChain: J: PDB Molecule: 4-oxalocrotonate tautomerase family enzyme - beta subunit;PDBTitle: kinetic and structural characterization of a heterohexamer 4-2 oxalocrotonate tautomerase from chloroflexus aurantiacus j-10-fl:3 implications for functional and structural diversity in the4 tautomerase superfamily
10 c3qwmA_
17.5
20
PDB header: signaling proteinChain: A: PDB Molecule: iq motif and sec7 domain-containing protein 1;PDBTitle: crystal structure of gep100, the plextrin homology domain of iq motif2 and sec7 domain-containing protein 1 isoform a
11 c3m8jA_
16.2
33
PDB header: transcriptionChain: A: PDB Molecule: focb protein;PDBTitle: crystal structure of e.coli focb at 1.4 a resolution
12 c1a87A_
15.8
25
PDB header: bacteriocinChain: A: PDB Molecule: colicin n;PDBTitle: colicin n
13 d1a87a_
15.8
25
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin14 c3h0dB_
14.2
22
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
15 d2ahra1
13.5
21
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: ProC C-terminal domain-like16 c2izzE_
9.7
19
PDB header: oxidoreductaseChain: E: PDB Molecule: pyrroline-5-carboxylate reductase 1;PDBTitle: crystal structure of human pyrroline-5-carboxylate2 reductase
17 c1s7cA_
9.4
22
PDB header: structural genomics, oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of mes buffer bound form of glyceraldehyde 3-2 phosphate dehydrogenase from escherichia coli
18 c2i5pO_
8.9
11
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase isoform 1 from k. marxianus
19 c3gtyS_
8.9
10
PDB header: chaperone/ribosomal proteinChain: S: PDB Molecule: 30s ribosomal protein s7;PDBTitle: promiscuous substrate recognition in folding and assembly activities2 of the trigger factor chaperone
20 c3h9eA_
8.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
21 c1hdgO_
not modelled
8.3
11
PDB header: oxidoreductase (aldehy(d)-nad(a))Chain: O: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
22 c3ci9B_
not modelled
8.3
21
PDB header: transcriptionChain: B: PDB Molecule: heat shock factor-binding protein 1;PDBTitle: crystal structure of the human hsbp1
23 c3b20R_
not modelled
8.2
9
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase (nadp+);PDBTitle: crystal structure analysis of dehydrogenase complexed with nad
24 c3hq4R_
not modelled
8.1
11
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
25 c3hjaB_
not modelled
8.0
11
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase from borrelia burgdorferi
26 c1ihxD_
not modelled
7.9
17
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of two d-glyceraldehyde-3-phosphate2 dehydrogenase complexes: a case of asymmetry
27 c2d2iO_
not modelled
7.9
11
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-2 phosphate dehydrogenase from synechococcus sp. complexed3 with nadp+
28 d1k6ka_
not modelled
7.6
16
Fold: Double Clp-N motifSuperfamily: Double Clp-N motifFamily: Double Clp-N motif29 c2x5kO_
not modelled
7.4
6
PDB header: oxidoreductaseChain: O: PDB Molecule: d-erythrose-4-phosphate dehydrogenase;PDBTitle: structure of an active site mutant of the d-erythrose-4-phosphate2 dehydrogenase from e. coli
30 c1qvrB_
not modelled
7.4
18
PDB header: chaperoneChain: B: PDB Molecule: clpb protein;PDBTitle: crystal structure analysis of clpb
31 c2ep7B_
not modelled
7.2
17
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
32 d1u94a2
not modelled
7.2
19
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain33 d2bida_
not modelled
7.1
9
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death34 c2pkrI_
not modelled
7.1
11
PDB header: oxidoreductaseChain: I: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase aor;PDBTitle: crystal structure of (a+cte)4 chimeric form of2 photosyntetic glyceraldehyde-3-phosphate dehydrogenase,3 complexed with nadp
35 c3docD_
not modelled
7.0
11
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of trka glyceraldehyde-3-phosphate2 dehydrogenase from brucella melitensis
36 c1rm4O_
not modelled
6.9
11
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of recombinant photosynthetic glyceraldehyde-3-2 phosphate dehydrogenase a4 isoform, complexed with nadp
37 c2fs1A_
not modelled
6.9
15
PDB header: protein bindingChain: A: PDB Molecule: psd-1;PDBTitle: solution structure of psd-1
38 d1xp8a2
not modelled
6.9
20
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain39 c2b4rQ_
not modelled
6.8
11
PDB header: oxidoreductaseChain: Q: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
40 c1i32D_
not modelled
6.8
13
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: leishmania mexicana glyceraldehyde-3-phosphate2 dehydrogenase in complex with inhibitors
41 c1cerC_
not modelled
6.6
6
PDB header: oxidoreductase (aldehyde(d)-nad(a))Chain: C: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: determinants of enzyme thermostability observed in the2 molecular structure of thermus aquaticus d-glyceraldehyde-3 3-phosphate dehydrogenase at 2.5 angstroms resolution
42 d1mo6a2
not modelled
6.5
16
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain43 d1ubea2
not modelled
6.4
16
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain44 c3cieC_
not modelled
6.3
17
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
45 c1obfO_
not modelled
6.2
11
PDB header: glycolytic pathwayChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.73 resolution.
46 d2c42a4
not modelled
6.1
18
Fold: Pyruvate-ferredoxin oxidoreductase, PFOR, domain IIISuperfamily: Pyruvate-ferredoxin oxidoreductase, PFOR, domain IIIFamily: Pyruvate-ferredoxin oxidoreductase, PFOR, domain III47 d1gjsa_
not modelled
6.1
23
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: GA module, an albumin-binding domain48 c2i88A_
not modelled
6.1
15
PDB header: membrane proteinChain: A: PDB Molecule: colicin-e1;PDBTitle: crystal structure of the channel-forming domain of colicin2 e1
49 d1v9va1
not modelled
6.1
17
Fold: Bromodomain-likeSuperfamily: MAST3 pre-PK domain-likeFamily: MAST3 pre-PK domain-like50 c3sthA_
not modelled
5.6
17
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 toxoplasma gondii
51 c2y69Z_
not modelled
5.5
17
PDB header: electron transportChain: Z: PDB Molecule: cytochrome c oxidase polypeptide 8h;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
52 c1g92A_
not modelled
5.4
67
PDB header: toxinChain: A: PDB Molecule: poneratoxin;PDBTitle: solution structure of poneratoxin
53 c2po3B_
not modelled
5.3
22
PDB header: transferaseChain: B: PDB Molecule: 4-dehydrase;PDBTitle: crystal structure analysis of desi in the presence of its2 tdp-sugar product
54 c2gd1P_
not modelled
5.1
11
PDB header: oxidoreductase(aldehyde(d)-nad(a))Chain: P: PDB Molecule: apo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: coenzyme-induced conformational changes in glyceraldehyde-3-2 phosphate dehydrogenase from bacillus stearothermophillus
55 d1hula_
not modelled
5.1
17
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Short-chain cytokines