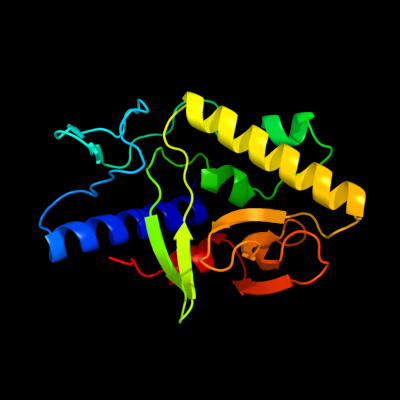
| 1 | c3ffvA_
|
|
 |
100.0 |
100 |
PDB header:protein binding
Chain: A: PDB Molecule:protein syd;
PDBTitle: crystal structure analysis of syd
|
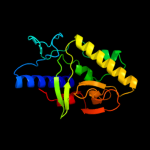
| 2 | c3d5pB_
|
|
 |
97.7 |
13 |
PDB header:gene regulation
Chain: B: PDB Molecule:putative glucan synthesis regulator of smi1/knr4 family;
PDBTitle: crystal structure of a putative glucan synthesis regulator of2 smi1/knr4 family (bf1740) from bacteroides fragilis nctc 9343 at 1.453 a resolution
|
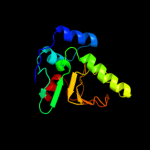
| 3 | d2prva1
|
|
 |
97.1 |
10 |
Fold:SMI1/KNR4-like
Superfamily:SMI1/KNR4-like
Family:SMI1/KNR4-like |
| 4 | d2icga1
|
|
 |
96.2 |
11 |
Fold:SMI1/KNR4-like
Superfamily:SMI1/KNR4-like
Family:SMI1/KNR4-like |
| 5 | d2paga1
|
|
 |
83.0 |
15 |
Fold:SMI1/KNR4-like
Superfamily:SMI1/KNR4-like
Family:SMI1/KNR4-like |
| 6 | c1hf9B_
|
|
 |
32.2 |
26 |
PDB header:atpase inhibitor
Chain: B: PDB Molecule:atpase inhibitor (mitochondrial);
PDBTitle: c-terminal coiled-coil domain from bovine if1
|
| 7 | c1z5sD_
|
|
 |
30.0 |
53 |
PDB header:ligase
Chain: D: PDB Molecule:ran-binding protein 2;
PDBTitle: crystal structure of a complex between ubc9, sumo-1,2 rangap1 and nup358/ranbp2
|
| 8 | c3o27B_
|
|
 |
28.5 |
5 |
PDB header:dna binding protein
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: the crystal structure of c68 from the hybrid virus-plasmid pssvx
|
| 9 | d2bv3a4
|
|
 |
23.6 |
17 |
Fold:Ferredoxin-like
Superfamily:EF-G C-terminal domain-like
Family:EF-G/eEF-2 domains III and V |
| 10 | c2wryA_
|
|
 |
20.1 |
22 |
PDB header:immune system
Chain: A: PDB Molecule:interleukin-1beta;
PDBTitle: crystal structure of chicken cytokine interleukin 1 beta
|
| 11 | d1e2wa2
|
|
 |
19.3 |
15 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
| 12 | d1ci3m2
|
|
 |
17.2 |
10 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
| 13 | c3kepA_
|
|
 |
16.4 |
17 |
PDB header:protein transport, rna binding protein
Chain: A: PDB Molecule:nucleoporin nup145;
PDBTitle: crystal structure of the autoproteolytic domain from the2 nuclear pore complex component nup145 from saccharomyces3 cerevisiae
|
| 14 | c1q2lA_
|
|
 |
14.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:protease iii;
PDBTitle: crystal structure of pitrilysin
|
| 15 | c2jbuB_
|
|
 |
13.9 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:insulin-degrading enzyme;
PDBTitle: crystal structure of human insulin degrading enzyme2 complexed with co-purified peptides.
|
| 16 | c3dx5A_
|
|
 |
12.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 17 | c2wk3A_
|
|
 |
11.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:insulin degrading enzyme;
PDBTitle: crystal structure of human insulin-degrading enzyme in2 complex with amyloid-beta (1-42)
|
| 18 | d1j83a_
|
|
 |
11.5 |
31 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Family 17 carbohydrate binding module, CBM17 |
| 19 | c3lgeF_
|
|
 |
11.4 |
11 |
PDB header:lyase/protein binding
Chain: F: PDB Molecule:sorting nexin-9;
PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
|
| 20 | c3lgeG_
|
|
 |
11.4 |
11 |
PDB header:lyase/protein binding
Chain: G: PDB Molecule:sorting nexin-9;
PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
|
| 21 | c3d22A_ |
|
not modelled |
11.2 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin h-type;
PDBTitle: crystal structure of a poplar thioredoxin h mutant,2 pttrxh4c61s
|
| 22 | d1ilr1_ |
|
not modelled |
10.9 |
30 |
Fold:beta-Trefoil
Superfamily:Cytokine
Family:Interleukin-1 (IL-1) |
| 23 | c1m45B_ |
|
not modelled |
10.6 |
25 |
PDB header:cell cycle protein
Chain: B: PDB Molecule:iq2 motif from myo2p, a class v myosin;
PDBTitle: crystal structure of mlc1p bound to iq2 of myo2p, a class v2 myosin
|
| 24 | c2j57J_ |
|
not modelled |
10.5 |
18 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:methylamine dehydrogenase heavy chain;
PDBTitle: x-ray reduced paraccocus denitrificans methylamine2 dehydrogenase n-quinol in complex with amicyanin.
|
| 25 | d2a6qb1 |
|
not modelled |
10.5 |
14 |
Fold:YefM-like
Superfamily:YefM-like
Family:YefM-like |
| 26 | d1st9a_ |
|
not modelled |
10.4 |
34 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 27 | d8i1ba_ |
|
not modelled |
10.1 |
30 |
Fold:beta-Trefoil
Superfamily:Cytokine
Family:Interleukin-1 (IL-1) |
| 28 | d2bg1a1 |
|
not modelled |
10.1 |
17 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 29 | c2kllA_ |
|
not modelled |
10.0 |
33 |
PDB header:cytokine
Chain: A: PDB Molecule:interleukin-33;
PDBTitle: solution structure of human interleukin-33
|
| 30 | c2hlwA_ |
|
not modelled |
9.2 |
15 |
PDB header:ligase, signaling protein
Chain: A: PDB Molecule:ubiquitin-conjugating enzyme e2 variant 1;
PDBTitle: solution structure of the human ubiquitin-conjugating2 enzyme variant uev1a
|
| 31 | c2a7oA_ |
|
not modelled |
8.9 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:huntingtin interacting protein b;
PDBTitle: solution structure of the hset2/hypb sri domain
|
| 32 | d1smya1 |
|
not modelled |
8.8 |
24 |
Fold:DCoH-like
Superfamily:RBP11-like subunits of RNA polymerase
Family:RNA polymerase alpha subunit dimerisation domain |
| 33 | d1md6a_ |
|
not modelled |
8.0 |
17 |
Fold:beta-Trefoil
Superfamily:Cytokine
Family:Interleukin-1 (IL-1) |
| 34 | d2v0ea1 |
|
not modelled |
7.8 |
16 |
Fold:GYF/BRK domain-like
Superfamily:BRK domain-like
Family:BRK domain-like |
| 35 | c3psiA_ |
|
not modelled |
7.6 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt6;
PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(239-1451)
|
| 36 | d1tu2b2 |
|
not modelled |
7.6 |
26 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
| 37 | c2nogA_ |
|
not modelled |
7.1 |
6 |
PDB header:dna binding protein
Chain: A: PDB Molecule:iswi protein;
PDBTitle: sant domain structure of xenopus remodeling factor iswi
|
| 38 | c3kh7A_ |
|
not modelled |
7.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbe;
PDBTitle: crystal structure of the periplasmic soluble domain of reduced ccmg2 from pseudomonas aeruginosa
|
| 39 | c3iu6A_ |
|
not modelled |
6.8 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:protein polybromo-1;
PDBTitle: crystal structure of the sixth bromodomain of human poly-bromodomain2 containing protein 1 (pb1)
|
| 40 | c2jciA_ |
|
not modelled |
6.8 |
17 |
PDB header:drug-binding protein
Chain: A: PDB Molecule:penicillin-binding protein 1b;
PDBTitle: structural insights into the catalytic mechanism and the2 role of streptococcus pneumoniae pbp1b
|
| 41 | c2j04C_ |
|
not modelled |
6.6 |
13 |
PDB header:transcription
Chain: C: PDB Molecule:hypothetical protein ypl007c;
PDBTitle: the tau60-tau91 subcomplex of yeast transcription factor2 iiic
|
| 42 | d1qy9a1 |
|
not modelled |
6.1 |
20 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 43 | d1ii2a2 |
|
not modelled |
6.0 |
32 |
Fold:PEP carboxykinase N-terminal domain
Superfamily:PEP carboxykinase N-terminal domain
Family:PEP carboxykinase N-terminal domain |
| 44 | c3uinD_ |
|
not modelled |
5.9 |
70 |
PDB header:ligase/isomerase/protein binding
Chain: D: PDB Molecule:e3 sumo-protein ligase ranbp2;
PDBTitle: complex between human rangap1-sumo2, ubc9 and the ir1 domain from2 ranbp2
|
| 45 | d1wb9a3 |
|
not modelled |
5.6 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:DNA repair protein MutS, domain II
Family:DNA repair protein MutS, domain II |
| 46 | d1zyma2 |
|
not modelled |
5.6 |
14 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system |
| 47 | d1yrva1 |
|
not modelled |
5.5 |
21 |
Fold:UBC-like
Superfamily:UBC-like
Family:UBC-related |
| 48 | c3b79A_ |
|
not modelled |
5.5 |
20 |
PDB header:nucleotide binding protein
Chain: A: PDB Molecule:toxin secretion atp-binding protein;
PDBTitle: crystal structure of the n-terminal peptidase c39 like2 domain of the toxin secretion atp-binding protein from3 vibrio parahaemolyticus
|
| 49 | c1n2dC_ |
|
not modelled |
5.5 |
25 |
PDB header:cell cycle
Chain: C: PDB Molecule:iq2 and iq3 motifs from myo2p, a class v myosin;
PDBTitle: ternary complex of mlc1p bound to iq2 and iq3 of myo2p, a2 class v myosin
|
| 50 | c2o3oD_ |
|
not modelled |
5.5 |
23 |
PDB header:signaling protein
Chain: D: PDB Molecule:yyci protein;
PDBTitle: crystal structure of the sensor histidine kinase regulator yyci from2 bacillus subtitlis
|
| 51 | d1tf5a1 |
|
not modelled |
5.4 |
22 |
Fold:Pre-protein crosslinking domain of SecA
Superfamily:Pre-protein crosslinking domain of SecA
Family:Pre-protein crosslinking domain of SecA |
| 52 | c1mzwB_ |
|
not modelled |
5.4 |
38 |
PDB header:isomerase
Chain: B: PDB Molecule:u4/u6 snrnp 60kda protein;
PDBTitle: crystal structure of a u4/u6 snrnp complex between human2 spliceosomal cyclophilin h and a u4/u6-60k peptide
|
| 53 | c2wadB_ |
|
not modelled |
5.3 |
17 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:penicillin-binding protein 2b;
PDBTitle: penicillin-binding protein 2b (pbp-2b) from streptococcus2 pneumoniae (strain 5204)
|
| 54 | d2olua2 |
|
not modelled |
5.2 |
8 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 55 | c1ezaA_ |
|
not modelled |
5.2 |
14 |
PDB header:phosphotransferase
Chain: A: PDB Molecule:enzyme i;
PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
|
| 56 | c1qy9B_ |
|
not modelled |
5.1 |
19 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein ydde;
PDBTitle: crystal structure of e. coli se-met protein ydde
|
| 57 | d2gw6a1 |
|
not modelled |
5.1 |
22 |
Fold:Restriction endonuclease-like
Superfamily:tRNA-intron endonuclease catalytic domain-like
Family:tRNA-intron endonuclease catalytic domain-like |
| 58 | d1s1qa_ |
|
not modelled |
5.1 |
19 |
Fold:UBC-like
Superfamily:UBC-like
Family:UEV domain |
| 59 | c3udiA_ |
|
not modelled |
5.1 |
17 |
PDB header:penicillin-binding protein/antibiotic
Chain: A: PDB Molecule:penicillin-binding protein 1a;
PDBTitle: crystal structure of acinetobacter baumannii pbp1a in complex with2 penicillin g
|