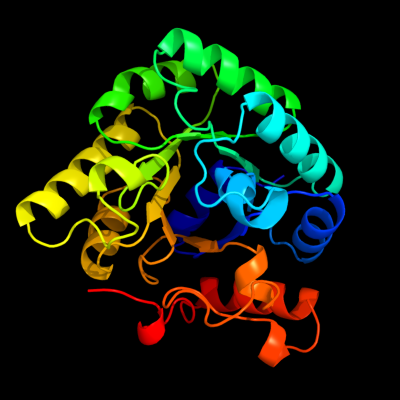
| 1 | d1m3ua_
|
|
 |
100.0 |
100 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
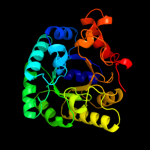
| 2 | d1o66a_
|
|
 |
100.0 |
51 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
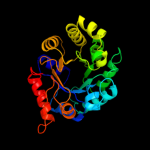
| 3 | d1oy0a_
|
|
 |
100.0 |
44 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
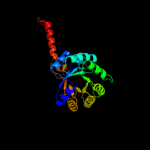
| 4 | c3ez4B_
|
|
 |
100.0 |
54 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 5 | c1zlpA_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 6 | d1muma_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 7 | d1ujqa_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 8 | c2ze3A_
|
|
 |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 9 | c3eooL_
|
|
 |
100.0 |
14 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 10 | c3ih1A_
|
|
 |
100.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 11 | c2hjpA_
|
|
 |
100.0 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 12 | c3b8iF_
|
|
 |
100.0 |
18 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 13 | c2qiwA_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 14 | c3fa4D_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 15 | c3lyeA_
|
|
 |
100.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 16 | d1s2wa_
|
|
 |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 17 | d1f61a_
|
|
 |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 18 | c3e5bB_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:isocitrate lyase;
PDBTitle: 2.4 a crystal structure of isocitrate lyase from brucella2 melitensis
|
| 19 | d1igwa_
|
|
 |
99.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 20 | d1dqua_
|
|
 |
99.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 21 | d1xcfa_ |
|
not modelled |
98.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 22 | d2p10a1 |
|
not modelled |
98.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Mll9387-like |
| 23 | c2p10D_ |
|
not modelled |
98.2 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 24 | d1gvfa_ |
|
not modelled |
97.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 25 | c3navB_ |
|
not modelled |
97.6 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 26 | c2z6jB_ |
|
not modelled |
97.5 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trans-2-enoyl-acp reductase ii;
PDBTitle: crystal structure of s. pneumoniae enoyl-acyl carrier2 protein reductase (fabk) in complex with an inhibitor
|
| 27 | d1qopa_ |
|
not modelled |
97.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 28 | d1xm3a_ |
|
not modelled |
97.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 29 | c2eq5D_ |
|
not modelled |
97.1 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:228aa long hypothetical hydantoin racemase;
PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
|
| 30 | c2gjlA_ |
|
not modelled |
97.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein pa1024;
PDBTitle: crystal structure of 2-nitropropane dioxygenase
|
| 31 | c3pm6B_ |
|
not modelled |
97.1 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-bisphosphate aldolase;
PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
|
| 32 | d1rd5a_ |
|
not modelled |
97.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 33 | c3igsB_ |
|
not modelled |
96.9 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
| 34 | c3q94B_ |
|
not modelled |
96.9 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase, class ii;
PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
|
| 35 | c3bw2A_ |
|
not modelled |
96.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-nitropropane dioxygenase;
PDBTitle: crystal structures and site-directed mutagenesis study of nitroalkane2 oxidase from streptomyces ansochromogenes
|
| 36 | d1rvga_ |
|
not modelled |
96.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 37 | c2iswB_ |
|
not modelled |
96.7 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-1,6-bisphosphate aldolase;
PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
|
| 38 | c2h90A_ |
|
not modelled |
96.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
| 39 | c2htmB_ |
|
not modelled |
96.6 |
18 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiazole biosynthesis protein thig;
PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
|
| 40 | d1jpma1 |
|
not modelled |
96.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 41 | d1vzwa1 |
|
not modelled |
96.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 42 | d1wv2a_ |
|
not modelled |
96.5 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 43 | c1zfjA_ |
|
not modelled |
96.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 44 | c1jvnB_ |
|
not modelled |
96.3 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 45 | c3elfA_ |
|
not modelled |
96.2 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: structural characterization of tetrameric mycobacterium tuberculosis2 fructose 1,6-bisphosphate aldolase - substrate binding and catalysis3 mechanism of a class iia bacterial aldolase
|
| 46 | c3lerA_ |
|
not modelled |
96.2 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 47 | c2rfgB_ |
|
not modelled |
96.1 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 48 | d1h5ya_ |
|
not modelled |
96.1 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 49 | c3hf3A_ |
|
not modelled |
96.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 50 | c3ffsC_ |
|
not modelled |
96.1 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: the crystal structure of cryptosporidium parvum inosine-5'-2 monophosphate dehydrogenase
|
| 51 | c3qm3C_ |
|
not modelled |
96.1 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: 1.85 angstrom resolution crystal structure of fructose-bisphosphate2 aldolase (fba) from campylobacter jejuni
|
| 52 | c3c52B_ |
|
not modelled |
96.0 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from2 helicobacter pylori in complex with3 phosphoglycolohydroxamic acid, a competitive inhibitor
|
| 53 | c2r94B_ |
|
not modelled |
96.0 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 54 | d1gwja_ |
|
not modelled |
96.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 55 | c3bo9B_ |
|
not modelled |
95.9 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nitroalkan dioxygenase;
PDBTitle: crystal structure of putative nitroalkan dioxygenase (tm0800) from2 thermotoga maritima at 2.71 a resolution
|
| 56 | c3daqB_ |
|
not modelled |
95.9 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 57 | c1jcnA_ |
|
not modelled |
95.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase i;
PDBTitle: binary complex of human type-i inosine monophosphate dehydrogenase2 with 6-cl-imp
|
| 58 | c2qjhH_ |
|
not modelled |
95.8 |
14 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
| 59 | d1dosa_ |
|
not modelled |
95.8 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 60 | d1vyra_ |
|
not modelled |
95.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 61 | c3n2xB_ |
|
not modelled |
95.7 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 62 | d1zfja1 |
|
not modelled |
95.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 63 | c2vlbC_ |
|
not modelled |
95.6 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 64 | c2y85D_ |
|
not modelled |
95.6 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:phosphoribosyl isomerase a;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
|
| 65 | d1jfla1 |
|
not modelled |
95.5 |
31 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 66 | c3b0vD_ |
|
not modelled |
95.5 |
18 |
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
|
| 67 | c3gr7A_ |
|
not modelled |
95.4 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 68 | c3gkaB_ |
|
not modelled |
95.4 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
| 69 | d1vjia_ |
|
not modelled |
95.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 70 | d1f76a_ |
|
not modelled |
95.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 71 | c2hmcA_ |
|
not modelled |
95.3 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
| 72 | c1ps9A_ |
|
not modelled |
95.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 73 | c2gq8A_ |
|
not modelled |
95.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: structure of sye1, an oye homologue from s. ondeidensis, in complex2 with p-hydroxyacetophenone
|
| 74 | c3q58A_ |
|
not modelled |
95.1 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
| 75 | d1rvka1 |
|
not modelled |
95.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 76 | d1ps9a1 |
|
not modelled |
95.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 77 | c2v9dB_ |
|
not modelled |
95.0 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 78 | d1ojxa_ |
|
not modelled |
95.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 79 | c3qvjB_ |
|
not modelled |
95.0 |
27 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
| 80 | d1ykwa1 |
|
not modelled |
94.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 81 | d1o5ka_ |
|
not modelled |
94.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 82 | d1ka9f_ |
|
not modelled |
94.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 83 | c3e96B_ |
|
not modelled |
94.6 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 84 | c1ypfB_ |
|
not modelled |
94.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gmp reductase;
PDBTitle: crystal structure of guac (ba5705) from bacillus anthracis at 1.8 a2 resolution
|
| 85 | d1y0ea_ |
|
not modelled |
94.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
| 86 | c2vc6A_ |
|
not modelled |
94.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 87 | c3nwrA_ |
|
not modelled |
94.3 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
| 88 | d1goxa_ |
|
not modelled |
94.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 89 | d1pv8a_ |
|
not modelled |
94.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 90 | c1vrdA_ |
|
not modelled |
94.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase (tm1347)2 from thermotoga maritima at 2.18 a resolution
|
| 91 | c2w6rA_ |
|
not modelled |
94.2 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:imidazole glycerol phosphate synthase subunit
PDBTitle: crystal structure of an artificial (ba)8-barrel protein2 designed from identical half barrels
|
| 92 | d1pvna1 |
|
not modelled |
94.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 93 | d1p4ca_ |
|
not modelled |
94.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 94 | c3fokH_ |
|
not modelled |
94.0 |
19 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:uncharacterized protein cgl0159;
PDBTitle: crystal structure of cgl0159 from corynebacterium2 glutamicum (brevibacterium flavum). northeast structural3 genomics target cgr115
|
| 95 | c3fluD_ |
|
not modelled |
94.0 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 96 | c3s5oA_ |
|
not modelled |
94.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 97 | c3noeA_ |
|
not modelled |
93.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 98 | c3obkH_ |
|
not modelled |
93.9 |
19 |
PDB header:lyase
Chain: H: PDB Molecule:delta-aminolevulinic acid dehydratase;
PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
|
| 99 | c2xecD_ |
|
not modelled |
93.9 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 100 | d1z41a1 |
|
not modelled |
93.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 101 | d1jr1a1 |
|
not modelled |
93.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 102 | c2vwtA_ |
|
not modelled |
93.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|
| 103 | d1tzza1 |
|
not modelled |
93.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 104 | c3ojcD_ |
|
not modelled |
93.7 |
29 |
PDB header:isomerase
Chain: D: PDB Molecule:putative aspartate/glutamate racemase;
PDBTitle: crystal structure of a putative asp/glu racemase from yersinia pestis
|
| 105 | c2jfzB_ |
|
not modelled |
93.7 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
|
| 106 | c2zskA_ |
|
not modelled |
93.6 |
26 |
PDB header:unknown function
Chain: A: PDB Molecule:226aa long hypothetical aspartate racemase;
PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
|
| 107 | c3jrkG_ |
|
not modelled |
93.6 |
17 |
PDB header:lyase
Chain: G: PDB Molecule:tagatose 1,6-diphosphate aldolase 2;
PDBTitle: a putative tagatose 1,6-diphosphate aldolase from streptococcus2 pyogenes
|
| 108 | c2ps2A_ |
|
not modelled |
93.6 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative mandelate racemase/muconate lactonizing
PDBTitle: crystal structure of putative mandelate racemase/muconate2 lactonizing enzyme from aspergillus oryzae
|
| 109 | c1telA_ |
|
not modelled |
93.6 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase, large subunit;
PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
|
| 110 | d2mnra1 |
|
not modelled |
93.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 111 | c3stgA_ |
|
not modelled |
93.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
|
| 112 | c1b74A_ |
|
not modelled |
93.5 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase from aquifex pyrophilus
|
| 113 | d1geqa_ |
|
not modelled |
93.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 114 | d2tpsa_ |
|
not modelled |
93.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 115 | d2c1ha1 |
|
not modelled |
93.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 116 | c2nuxB_ |
|
not modelled |
93.3 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
| 117 | c3fkkA_ |
|
not modelled |
93.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
| 118 | c3s81A_ |
|
not modelled |
93.2 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
|
| 119 | c3pueA_ |
|
not modelled |
93.2 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 120 | d1geha1 |
|
not modelled |
93.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |