1 c3rfzB_
100.0
12
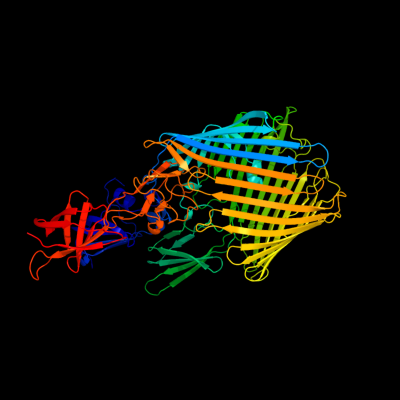
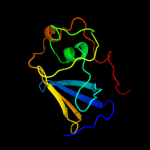
PDB header: cell adhesion/transport/chaperoneChain: B: PDB Molecule: outer membrane usher protein, type 1 fimbrial synthesis;PDBTitle: crystal structure of the fimd usher bound to its cognate fimc:fimh2 substrate
2 c3ohnA_
100.0
12
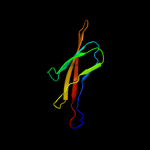
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane usher protein fimd;PDBTitle: crystal structure of the fimd translocation domain
3 c2vqiA_
100.0
15
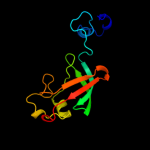
PDB header: transportChain: A: PDB Molecule: outer membrane usher protein papc;PDBTitle: structure of the p pilus usher (papc) translocation pore
4 d1zdva1
99.9
18
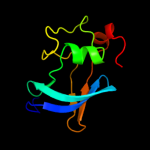
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain5 c3fcgB_
99.8
18
PDB header: membrane protein, protein transportChain: B: PDB Molecule: f1 capsule-anchoring protein;PDBTitle: crystal structure analysis of the middle domain of the2 caf1a usher
6 d3bwud1
99.7
16
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain7 d1zdxa1
99.7
19
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain8 c3l48B_
99.4
16
PDB header: transport proteinChain: B: PDB Molecule: outer membrane usher protein papc;PDBTitle: crystal structure of the c-terminal domain of the papc usher
9 c2xetB_
99.3
10
PDB header: transport proteinChain: B: PDB Molecule: f1 capsule-anchoring protein;PDBTitle: conserved hydrophobic clusters on the surface of the caf1a usher2 c-terminal domain are important for f1 antigen assembly
10 c1uwyA_
95.5
17
PDB header: hydrolaseChain: A: PDB Molecule: carboxypeptidase m;PDBTitle: crystal structure of human carboxypeptidase m
11 d1h8la1
95.3
15
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain12 d1uwya1
94.8
21
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain13 c2nsmA_
94.8
17
PDB header: hydrolaseChain: A: PDB Molecule: carboxypeptidase n catalytic chain;PDBTitle: crystal structure of the human carboxypeptidase n (kininase i)2 catalytic domain
14 c1h8lA_
94.6
15
PDB header: carboxypeptidaseChain: A: PDB Molecule: carboxypeptidase gp180 residues 503-882;PDBTitle: duck carboxypeptidase d domain ii in complex with gemsa
15 c3mn8A_
94.1
18
PDB header: hydrolaseChain: A: PDB Molecule: lp15968p;PDBTitle: structure of drosophila melanogaster carboxypeptidase d isoform 1b2 short
16 d2burb1
91.8
43
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase17 d3pccm_
91.5
27
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase18 c2azqA_
89.7
23
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure of catechol 1,2-dioxygenase from pseudomonas arvilla2 c-1
19 c3n9tA_
88.9
23
PDB header: oxidoreductaseChain: A: PDB Molecule: pnpc;PDBTitle: cryatal structure of hydroxyquinol 1,2-dioxygenase from pseudomonas2 putida dll-e4
20 c3pdgA_
87.6
13
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
21 c3hj8A_
not modelled
87.0
30
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure determination of catechol 1,2-dioxygenase from2 rhodococcus opacus 1cp in complex with 4-chlorocatechol
22 d1nkga1
not modelled
87.0
15
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Rhamnogalacturonase B, RhgB, middle domain23 d1s9aa_
not modelled
86.8
25
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase24 c1tmxA_
not modelled
86.5
20
PDB header: oxidoreductaseChain: A: PDB Molecule: hydroxyquinol 1,2-dioxygenase;PDBTitle: crystal structure of hydroxyquinol 1,2-dioxygenase from2 nocardioides simplex 3e
25 c3pe9B_
not modelled
86.4
12
PDB header: unknown functionChain: B: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
26 c3e8vA_
not modelled
86.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: possible transglutaminase-family protein;PDBTitle: crystal structure of a possible transglutaminase-family2 protein proteolytic fragment from bacteroides fragilis
27 c2xsuA_
not modelled
86.1
18
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2 dioxygenase;PDBTitle: crystal structure of the a72g mutant of acinetobacter2 radioresistens catechol 1,2 dioxygenase
28 c2boyC_
not modelled
82.0
26
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-chlorocatechol 1,2-dioxygenase;PDBTitle: crystal structure of 3-chlorocatechol 1,2-dioxygenase from2 rhodococcus opacus 1cp
29 d1dmha_
not modelled
80.6
32
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase30 c3pe9D_
not modelled
80.2
15
PDB header: unknown functionChain: D: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
31 c2vnvC_
not modelled
71.4
14
PDB header: sugar-binding proteinChain: C: PDB Molecule: bcla;PDBTitle: crystal structure of bcla lectin from burkholderia2 cenocepacia in complex with alpha-methyl-mannoside at 1.73 angstrom resolution
32 d3pcca_
not modelled
64.3
23
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase33 d2q9oa2
not modelled
62.1
14
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins34 c1d2pA_
not modelled
55.4
15
PDB header: structural proteinChain: A: PDB Molecule: collagen adhesin;PDBTitle: crystal structure of two b repeat units (b1b2) of the2 collagen binding protein (cna) of staphylococcus aureus
35 d1w0na_
not modelled
55.2
15
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 36 carbohydrate binding module, CBM3636 c3pe9A_
not modelled
52.1
14
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
37 c3pe9C_
not modelled
52.1
14
PDB header: unknown functionChain: C: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
38 d2bura1
not modelled
51.8
20
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase39 d1kyaa2
not modelled
49.8
15
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins40 d1hfua2
not modelled
48.7
16
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins41 c2w87B_
not modelled
44.4
13
PDB header: hydrolaseChain: B: PDB Molecule: esterase d;PDBTitle: xyl-cbm35 in complex with glucuronic acid containing2 disaccharide.
42 d1aoza2
not modelled
42.1
17
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins43 d1v10a2
not modelled
41.8
19
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins44 d1cwva2
not modelled
41.0
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments45 c2r32A_
not modelled
40.0
19
PDB header: immune systemChain: A: PDB Molecule: gcn4-pii/tumor necrosis factor ligandPDBTitle: crystal structure of human gitrl variant
46 d1cwva1
not modelled
38.9
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments47 d1gyca2
not modelled
36.9
15
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins48 d1edqa1
not modelled
35.4
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes49 c3d33B_
not modelled
32.9
13
PDB header: unknown functionChain: B: PDB Molecule: domain of unknown function with an immunoglobulin-likePDBTitle: crystal structure of a duf3244 family protein with an immunoglobulin-2 like beta-sandwich fold (bvu_0276) from bacteroides vulgatus atcc3 8482 at 1.70 a resolution
50 c2x5pA_
not modelled
25.4
19
PDB header: protein bindingChain: A: PDB Molecule: fibronectin binding protein;PDBTitle: crystal structure of the streptococcus pyogenes fibronectin binding2 protein fbab-b
51 c3ottB_
not modelled
24.6
15
PDB header: transcriptionChain: B: PDB Molecule: two-component system sensor histidine kinase;PDBTitle: crystal structure of the extracellular domain of the putative one2 component system bt4673 from b. thetaiotaomicron
52 c2crvA_
not modelled
24.6
19
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-2;PDBTitle: solution structure of c-terminal domain of mitochondrial2 translational initiationfactor 2
53 c3c12A_
not modelled
21.7
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: flagellar protein;PDBTitle: crystal structure of flgd from xanthomonas campestris:2 insights into the hook capping essential for flagellar3 assembly
54 c3b9eA_
not modelled
21.5
13
PDB header: hydrolaseChain: A: PDB Molecule: chitinase a;PDBTitle: crystal structure of inactive mutant e315m chitinase a from2 vibrio harveyi
55 c1bprA_
not modelled
21.4
21
PDB header: molecular chaperoneChain: A: PDB Molecule: dnak;PDBTitle: nmr structure of the substrate binding domain of dnak,2 minimized average structure
56 c3uotB_
not modelled
20.4
12
PDB header: cell cycleChain: B: PDB Molecule: mediator of dna damage checkpoint protein 1;PDBTitle: crystal structure of mdc1 fha domain in complex with a phosphorylated2 peptide from the mdc1 n-terminus
57 d1f00i1
not modelled
20.2
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments58 c1u00A_
not modelled
20.1
12
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein hsca;PDBTitle: hsca substrate binding domain complexed with the iscu2 recognition peptide elppvkihc
59 d1u00a2
not modelled
19.6
19
Fold: Heat shock protein 70kD (HSP70), peptide-binding domainSuperfamily: Heat shock protein 70kD (HSP70), peptide-binding domainFamily: Heat shock protein 70kD (HSP70), peptide-binding domain60 c3osvC_
not modelled
19.6
18
PDB header: structural proteinChain: C: PDB Molecule: flagellar basal-body rod modification protein flgd;PDBTitle: the crytsal structure of flgd from p. aeruginosa
61 c3n8eA_
not modelled
19.0
17
PDB header: chaperoneChain: A: PDB Molecule: stress-70 protein, mitochondrial;PDBTitle: substrate binding domain of the human heat shock 70kda protein 92 (mortalin)
62 c3pddA_
not modelled
18.2
11
PDB header: unknown functionChain: A: PDB Molecule: glycoside hydrolase, family 9;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
63 c3dpqE_
not modelled
16.3
21
PDB header: chaperone, peptide binding proteinChain: E: PDB Molecule: chaperone protein dnak;PDBTitle: crystal structure of the substrate binding domain of e.2 coli dnak in complex with a long pyrrhocoricin-derived3 inhibitor peptide (form b)
64 d1thqa_
not modelled
15.9
6
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane enzyme PagP65 d1cwva3
not modelled
15.8
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments66 d1ulva2
not modelled
15.2
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes67 d1o75a2
not modelled
14.0
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Tp47 lipoprotein, middle and C-terminal domainsFamily: Tp47 lipoprotein, middle and C-terminal domains68 d1d1na_
not modelled
13.6
17
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors69 c1cwvA_
not modelled
12.1
17
PDB header: structural proteinChain: A: PDB Molecule: invasin;PDBTitle: crystal structure of invasin: a bacterial integrin-binding protein
70 c3dqgC_
not modelled
11.4
16
PDB header: chaperoneChain: C: PDB Molecule: heat shock 70 kda protein f;PDBTitle: peptide-binding domain of heat shock 70 kda protein f, mitochondrial2 precursor, from caenorhabditis elegans.
71 d1yuwa1
not modelled
11.3
17
Fold: Heat shock protein 70kD (HSP70), peptide-binding domainSuperfamily: Heat shock protein 70kD (HSP70), peptide-binding domainFamily: Heat shock protein 70kD (HSP70), peptide-binding domain72 d1zata1
not modelled
10.7
18
Fold: L,D-transpeptidase catalytic domain-likeSuperfamily: L,D-transpeptidase catalytic domain-likeFamily: L,D-transpeptidase catalytic domain-like73 d2o14a1
not modelled
10.1
40
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: YxiM N-terminal domain-like74 d2i02a1
not modelled
9.9
8
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like75 c2hklB_
not modelled
9.5
20
PDB header: transferaseChain: B: PDB Molecule: l,d-transpeptidase;PDBTitle: crystal structure of enterococcus faecium l,d-2 transpeptidase c442s mutant
76 c2qkiA_
not modelled
9.1
11
PDB header: immune system/hydrolase inhibitorChain: A: PDB Molecule: complement c3;PDBTitle: human c3c in complex with the inhibitor compstatin
77 c1motA_
not modelled
9.1
14
PDB header: membrane proteinChain: A: PDB Molecule: glycine receptor alpha-1 chain;PDBTitle: nmr structure of extended second transmembrane domain of2 glycine receptor alpha1 subunit in sds micelles
78 c2xr4A_
not modelled
9.0
19
PDB header: sugar binding proteinChain: A: PDB Molecule: lectin;PDBTitle: c-terminal domain of bc2l-c lectin from burkholderia cenocepacia
79 c2op6A_
not modelled
8.9
21
PDB header: peptide binding proteinChain: A: PDB Molecule: heat shock 70 kda protein d;PDBTitle: peptide-binding domain of heat shock 70 kda protein d precursor from2 c.elegans
80 c1cvrA_
not modelled
8.6
12
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: gingipain r;PDBTitle: crystal structure of the arg specific cysteine proteinase gingipain r2 (rgpb)
81 c2kppA_
not modelled
8.3
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin0431 protein;PDBTitle: solution nmr structure of lin0431 protein from listeria innocua.2 northeast structural genomics consortium target lkr112
82 c2w3jA_
not modelled
8.3
10
PDB header: sugar-binding proteinChain: A: PDB Molecule: carbohydrate binding module;PDBTitle: structure of a family 35 carbohydrate binding module from2 an environmental isolate
83 d2je8a4
not modelled
8.2
10
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain84 d1v8ha1
not modelled
7.9
4
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: SoxZ-like85 c3k1dA_
not modelled
7.9
22
PDB header: transferaseChain: A: PDB Molecule: 1,4-alpha-glucan-branching enzyme;PDBTitle: crystal structure of glycogen branching enzyme synonym: 1,4-alpha-d-2 glucan:1,4-alpha-d-glucan 6-glucosyl-transferase from mycobacterium3 tuberculosis h37rv
86 c1ug9A_
not modelled
7.6
16
PDB header: hydrolaseChain: A: PDB Molecule: glucodextranase;PDBTitle: crystal structure of glucodextranase from arthrobacter globiformis i42
87 c1vryA_
not modelled
7.4
8
PDB header: membrane proteinChain: A: PDB Molecule: glycine receptor alpha-1 chain;PDBTitle: second and third transmembrane domains of the alpha-12 subunit of human glycine receptor
88 d1yq2a3
not modelled
7.4
21
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain89 d1c8ua1
not modelled
7.4
13
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: Acyl-CoA thioesterase90 d3btaa1
not modelled
7.3
15
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Clostridium neurotoxins, the second last domain91 c2nn6I_
not modelled
7.2
12
PDB header: hydrolase/transferaseChain: I: PDB Molecule: 3'-5' exoribonuclease csl4 homolog;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
92 d1dkza2
not modelled
7.2
20
Fold: Heat shock protein 70kD (HSP70), peptide-binding domainSuperfamily: Heat shock protein 70kD (HSP70), peptide-binding domainFamily: Heat shock protein 70kD (HSP70), peptide-binding domain93 c3dnoC_
not modelled
7.2
13
PDB header: viral proteinChain: C: PDB Molecule: hiv-1 envelope glycoprotein gp120;PDBTitle: molecular structure for the hiv-1 gp120 trimer in the cd4-2 bound state
94 c3pvmB_
not modelled
7.2
13
PDB header: immune systemChain: B: PDB Molecule: cobra venom factor;PDBTitle: structure of complement c5 in complex with cvf
95 c2w1wB_
not modelled
7.0
15
PDB header: hydrolaseChain: B: PDB Molecule: lipolytic enzyme, g-d-s-l;PDBTitle: native structure of a family 35 carbohydrate binding module2 from clostridium thermocellum
96 d1qbea_
not modelled
6.8
12
Fold: RNA bacteriophage capsid proteinSuperfamily: RNA bacteriophage capsid proteinFamily: RNA bacteriophage capsid protein97 c2xwxB_
not modelled
6.8
11
PDB header: chitin-binding proteinChain: B: PDB Molecule: glcnac-binding protein a;PDBTitle: vibrio cholerae colonization factor gbpa crystal structure
98 c3kptA_
not modelled
6.7
20
PDB header: cell adhesionChain: A: PDB Molecule: collagen adhesion protein;PDBTitle: crystal structure of bcpa, the major pilin subunit of2 bacillus cereus
99 d2vmha1
not modelled
6.6
12
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: NPCBM-like