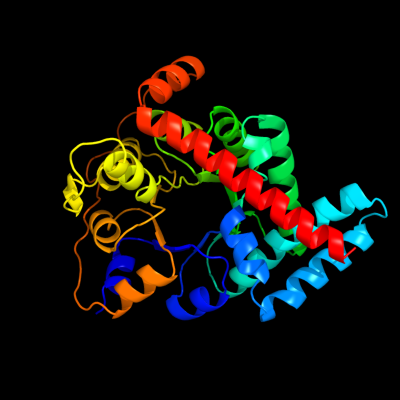
| 1 | d1onra_
|
|
 |
100.0 |
100 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
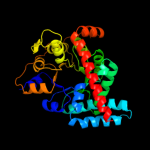
| 2 | d1f05a_
|
|
 |
100.0 |
60 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
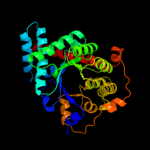
| 3 | d2e1da1
|
|
 |
100.0 |
60 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 4 | c3m16A_
|
|
 |
100.0 |
61 |
PDB header:transferase
Chain: A: PDB Molecule:transaldolase;
PDBTitle: structure of a transaldolase from oleispira antarctica
|
| 5 | c3igxA_
|
|
 |
100.0 |
48 |
PDB header:transferase
Chain: A: PDB Molecule:transaldolase;
PDBTitle: 1.85 angstrom resolution crystal structure of transaldolase b (tala)2 from francisella tularensis.
|
| 6 | c3hjzA_
|
|
 |
100.0 |
53 |
PDB header:transferase
Chain: A: PDB Molecule:transaldolase b;
PDBTitle: the structure of an aldolase from prochlorococcus marinus
|
| 7 | c3cq0B_
|
|
 |
100.0 |
53 |
PDB header:transferase
Chain: B: PDB Molecule:putative transaldolase ygr043c;
PDBTitle: crystal structure of tal2_yeast
|
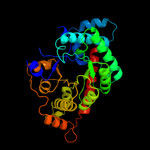
| 8 | c3clmA_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:transaldolase;
PDBTitle: crystal structure of transaldolase (yp_208650.1) from neisseria2 gonorrhoeae fa 1090 at 1.14 a resolution
|
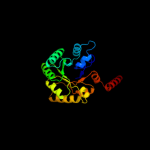
| 9 | c3s1vD_
|
|
 |
100.0 |
37 |
PDB header:transferase
Chain: D: PDB Molecule:probable transaldolase;
PDBTitle: transaldolase from thermoplasma acidophilum in complex with d-fructose2 6-phosphate schiff-base intermediate
|
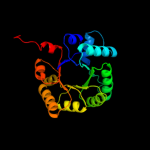
| 10 | d1wx0a1
|
|
 |
100.0 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 11 | d1vpxa_
|
|
 |
100.0 |
39 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
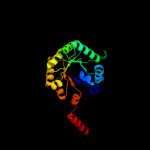
| 12 | d1l6wa_
|
|
 |
100.0 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 13 | d1xm3a_
|
|
 |
96.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 14 | c2htmB_
|
|
 |
95.0 |
18 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiazole biosynthesis protein thig;
PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
|
| 15 | c2p10D_
|
|
 |
92.5 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 16 | d1o4ua1
|
|
 |
87.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 17 | c1o4uA_
|
|
 |
85.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:type ii quinolic acid phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate nucleotide pyrophosphorylase2 (tm1645) from thermotoga maritima at 2.50 a resolution
|
| 18 | d2p10a1
|
|
 |
84.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Mll9387-like |
| 19 | c3labA_
|
|
 |
83.8 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate)
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
| 20 | c3gk0H_
|
|
 |
80.6 |
28 |
PDB header:transferase
Chain: H: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic2 protein from burkholderia pseudomallei
|
| 21 | d1ea0a2 |
|
not modelled |
77.6 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 22 | d1m5wa_ |
|
not modelled |
75.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Pyridoxine 5'-phosphate synthase
Family:Pyridoxine 5'-phosphate synthase |
| 23 | d1o66a_ |
|
not modelled |
72.1 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 24 | c3ez4B_ |
|
not modelled |
70.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 25 | c2jbmA_ |
|
not modelled |
70.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: qprtase structure from human
|
| 26 | c3o6cA_ |
|
not modelled |
69.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: pyridoxal phosphate biosynthetic protein pdxj from campylobacter2 jejuni
|
| 27 | d1nkua_ |
|
not modelled |
65.8 |
23 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:3-Methyladenine DNA glycosylase I (Tag) |
| 28 | d1wa3a1 |
|
not modelled |
64.3 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 29 | d1nu5a1 |
|
not modelled |
64.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 30 | d1ofda2 |
|
not modelled |
64.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 31 | d1cjca2 |
|
not modelled |
62.0 |
19 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 32 | d1kbia1 |
|
not modelled |
60.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 33 | c2yw3E_ |
|
not modelled |
60.8 |
25 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
| 34 | d1wbha1 |
|
not modelled |
59.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 35 | c3oa3A_ |
|
not modelled |
59.4 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 coccidioides immitis
|
| 36 | c2p0iA_ |
|
not modelled |
59.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:l-rhamnonate dehydratase;
PDBTitle: crystal structure of l-rhamnonate dehydratase from gibberella zeae
|
| 37 | c2hxtA_ |
|
not modelled |
58.1 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:l-fuconate dehydratase;
PDBTitle: crystal structure of l-fuconate dehydratase from xanthomonas2 campestris liganded with mg++ and d-erythronohydroxamate
|
| 38 | d1mzha_ |
|
not modelled |
56.1 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 39 | c2jg6A_ |
|
not modelled |
55.7 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna-3-methyladenine glycosidase;
PDBTitle: crystal structure of a 3-methyladenine dna glycosylase i2 from staphylococcus aureus
|
| 40 | c2b7pA_ |
|
not modelled |
54.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori
|
| 41 | d1wv2a_ |
|
not modelled |
52.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 42 | c1kbiB_ |
|
not modelled |
52.3 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome b2;
PDBTitle: crystallographic study of the recombinant flavin-binding domain of2 baker's yeast flavocytochrome b2: comparison with the intact wild-3 type enzyme
|
| 43 | c3ffsC_ |
|
not modelled |
50.8 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: the crystal structure of cryptosporidium parvum inosine-5'-2 monophosphate dehydrogenase
|
| 44 | d1pv8a_ |
|
not modelled |
50.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 45 | d1iyna_ |
|
not modelled |
50.5 |
21 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:CCP-like |
| 46 | c3pajA_ |
|
not modelled |
49.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase, carboxylating;
PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
|
| 47 | c1zfjA_ |
|
not modelled |
49.6 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 48 | c1lm1A_ |
|
not modelled |
49.4 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin-dependent glutamate synthase;
PDBTitle: structural studies on the synchronization of catalytic centers in2 glutamate synthase: native enzyme
|
| 49 | c1cjcA_ |
|
not modelled |
46.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (adrenodoxin reductase);
PDBTitle: structure of adrenodoxin reductase of mitochondrial p4502 systems
|
| 50 | c3cxoA_ |
|
not modelled |
43.6 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:putative galactonate dehydratase;
PDBTitle: crystal structure of l-rhamnonate dehydratase from2 salmonella typhimurium complexed with mg and 3-deoxy-l-3 rhamnonate
|
| 51 | d2euta1 |
|
not modelled |
43.5 |
23 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:CCP-like |
| 52 | c3rcyC_ |
|
not modelled |
42.7 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:mandelate racemase/muconate lactonizing enzyme-like
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme-2 like protein from roseovarius sp. tm1035
|
| 53 | c2v3aA_ |
|
not modelled |
40.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rubredoxin reductase;
PDBTitle: crystal structure of rubredoxin reductase from pseudomonas2 aeruginosa.
|
| 54 | d1j5ta_ |
|
not modelled |
40.1 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 55 | d1vc4a_ |
|
not modelled |
40.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 56 | d1vhna_ |
|
not modelled |
39.9 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 57 | d1eepa_ |
|
not modelled |
39.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 58 | d1d7ya2 |
|
not modelled |
39.6 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 59 | c2oz3F_ |
|
not modelled |
39.6 |
15 |
PDB header:lyase
Chain: F: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of l-rhamnonate dehydratase from azotobacter2 vinelandii
|
| 60 | d1vhca_ |
|
not modelled |
39.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 61 | c3l0gD_ |
|
not modelled |
37.1 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
|
| 62 | c3ef6A_ |
|
not modelled |
37.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:toluene 1,2-dioxygenase system ferredoxin--nad(+)
PDBTitle: crystal structure of toluene 2,3-dioxygenase reductase
|
| 63 | d1bd3a_ |
|
not modelled |
37.0 |
10 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 64 | d1p0ka_ |
|
not modelled |
36.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 65 | c3dg7B_ |
|
not modelled |
36.2 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:muconate cycloisomerase;
PDBTitle: crystal structure of muconate lactonizing enzyme from mucobacterium2 smegmatis complexed with muconolactone
|
| 66 | d2e39a1 |
|
not modelled |
35.8 |
18 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:CCP-like |
| 67 | d1mxsa_ |
|
not modelled |
35.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 68 | c3n4eA_ |
|
not modelled |
35.4 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme, c-terminal
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing protein2 from paracoccus denitrificans pd1222
|
| 69 | c3llvA_ |
|
not modelled |
35.0 |
10 |
PDB header:nad(p) binding protein
Chain: A: PDB Molecule:exopolyphosphatase-related protein;
PDBTitle: the crystal structure of the nad(p)-binding domain of an2 exopolyphosphatase-related protein from archaeoglobus fulgidus to3 1.7a
|
| 70 | c3gr7A_ |
|
not modelled |
34.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 71 | c2gr2A_ |
|
not modelled |
34.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
| 72 | d1qpoa1 |
|
not modelled |
33.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 73 | c2v82A_ |
|
not modelled |
33.1 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
| 74 | c2qq6B_ |
|
not modelled |
32.2 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:mandelate racemase/muconate lactonizing enzyme-
PDBTitle: crystal structure of mandelate racemase/muconate2 lactonizing enzyme-like protein from rubrobacter3 xylanophilus dsm 9941
|
| 75 | c3gkaB_ |
|
not modelled |
32.2 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
| 76 | d1vyra_ |
|
not modelled |
32.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 77 | c2pp1C_ |
|
not modelled |
31.5 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:l-talarate/galactarate dehydratase;
PDBTitle: crystal structure of l-talarate/galactarate dehydratase from2 salmonella typhimurium lt2 liganded with mg and l-lyxarohydroxamate
|
| 78 | c1x1oC_ |
|
not modelled |
30.7 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
|
| 79 | d2gdqa1 |
|
not modelled |
30.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 80 | c2ox4E_ |
|
not modelled |
29.8 |
17 |
PDB header:isomerase
Chain: E: PDB Molecule:putative mandelate racemase;
PDBTitle: crystal structure of putative dehydratase from zymomonas mobilis zm4
|
| 81 | d1z41a1 |
|
not modelled |
29.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 82 | c3hyxC_ |
|
not modelled |
29.0 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:sulfide-quinone reductase;
PDBTitle: 3-d x-ray structure of the sulfide:quinone oxidoreductase from aquifex2 aeolicus in complex with aurachin c
|
| 83 | c3px5A_ |
|
not modelled |
28.5 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:enzyme of enolase superfamily;
PDBTitle: structure of efi enolase target en500555, a putative dipeptide2 epimerase: apo structure
|
| 84 | c3q3uA_ |
|
not modelled |
28.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lignin peroxidase;
PDBTitle: trametes cervina lignin peroxidase
|
| 85 | c2vdcF_ |
|
not modelled |
28.1 |
24 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glutamate synthase [nadph] large chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 86 | c3t6cB_ |
|
not modelled |
27.5 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:putative mand family dehydratase;
PDBTitle: crystal structure of an enolase from pantoea ananatis (efi target efi-2 501676) with bound d-gluconate and mg
|
| 87 | c3riwA_ |
|
not modelled |
27.3 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ascorbate peroxidase;
PDBTitle: the crystal structure of leishmania major peroxidase mutant c197t
|
| 88 | d2dw4a2 |
|
not modelled |
27.1 |
27 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 89 | d1n7ka_ |
|
not modelled |
27.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 90 | d1goxa_ |
|
not modelled |
27.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 91 | c1tv5A_ |
|
not modelled |
26.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydroorotate dehydrogenase homolog, mitochondrial;
PDBTitle: plasmodium falciparum dihydroorotate dehydrogenase with a bound2 inhibitor
|
| 92 | d1tv5a1 |
|
not modelled |
26.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 93 | d1i5za1 |
|
not modelled |
26.7 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 94 | c1qpoA_ |
|
not modelled |
26.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:quinolinate acid phosphoribosyl transferase;
PDBTitle: quinolinate phosphoribosyl transferase (qaprtase) apo-enzyme from2 mycobacterium tuberculosis
|
| 95 | c3lxdA_ |
|
not modelled |
26.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: crystal structure of ferredoxin reductase arr from novosphingobium2 aromaticivorans
|
| 96 | d1f61a_ |
|
not modelled |
26.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 97 | c2rusB_ |
|
not modelled |
26.0 |
19 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:rubisco (ribulose-1,5-bisphosphate
PDBTitle: crystal structure of the ternary complex of ribulose-1,5-2 bisphosphate carboxylase, mg(ii), and activator co2 at 2.3-3 angstroms resolution
|
| 98 | c3fg2P_ |
|
not modelled |
25.8 |
25 |
PDB header:oxidoreductase
Chain: P: PDB Molecule:putative rubredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase for the cyp199a2 system from2 rhodopseudomonas palustris
|
| 99 | c1qapA_ |
|
not modelled |
25.8 |
12 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:quinolinic acid phosphoribosyltransferase;
PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
|
| 100 | c3fwzA_ |
|
not modelled |
25.6 |
10 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ybal;
PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
|
| 101 | c3h12B_ |
|
not modelled |
25.3 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:mandelate racemase;
PDBTitle: crystal structure of putative mandelate racemase from bordetella2 bronchiseptica rb50
|
| 102 | d1vkfa_ |
|
not modelled |
25.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:GlpP-like
Family:GlpP-like |
| 103 | c3bw2A_ |
|
not modelled |
25.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-nitropropane dioxygenase;
PDBTitle: crystal structures and site-directed mutagenesis study of nitroalkane2 oxidase from streptomyces ansochromogenes
|
| 104 | c1yqzA_ |
|
not modelled |
25.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme a disulfide reductase;
PDBTitle: structure of coenzyme a-disulfide reductase from2 staphylococcus aureus refined at 1.54 angstrom resolution
|
| 105 | c2ehjA_ |
|
not modelled |
24.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: structure of uracil phosphoribosyl transferase
|
| 106 | c1q1wA_ |
|
not modelled |
24.2 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putidaredoxin reductase;
PDBTitle: crystal structure of putidaredoxin reductase from2 pseudomonas putida
|
| 107 | d1nhpa2 |
|
not modelled |
24.1 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 108 | d1itza3 |
|
not modelled |
24.0 |
27 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Transketolase C-terminal domain-like |
| 109 | c2zrvC_ |
|
not modelled |
23.8 |
17 |
PDB header:isomerase
Chain: C: PDB Molecule:isopentenyl-diphosphate delta-isomerase;
PDBTitle: crystal structure of sulfolobus shibatae isopentenyl2 diphosphate isomerase in complex with reduced fmn.
|
| 110 | d1tb3a1 |
|
not modelled |
23.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 111 | c3ivuB_ |
|
not modelled |
23.8 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 112 | d1uuma_ |
|
not modelled |
23.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 113 | d1o5oa_ |
|
not modelled |
23.7 |
16 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 114 | c3sqsA_ |
|
not modelled |
23.5 |
7 |
PDB header:isomerase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing protein;
PDBTitle: crystal structure of a putative mandelate racemase/muconate2 lactonizing protein from dinoroseobacter shibae dfl 12
|
| 115 | c1rcxH_ |
|
not modelled |
23.4 |
21 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
| 116 | d1p4ca_ |
|
not modelled |
23.1 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 117 | d2chra1 |
|
not modelled |
22.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 118 | d1v9sa1 |
|
not modelled |
22.7 |
14 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 119 | c1me9A_ |
|
not modelled |
22.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh) from2 tritrichomonas foetus with imp bound
|
| 120 | d2gl5a1 |
|
not modelled |
21.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |