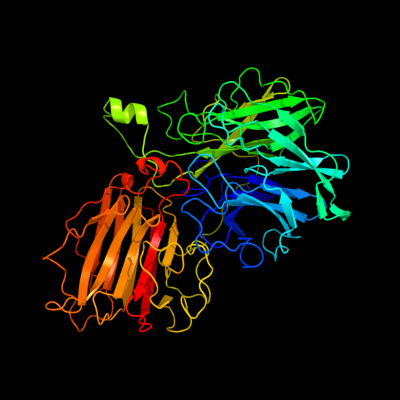
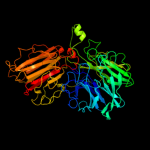
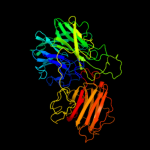
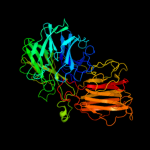
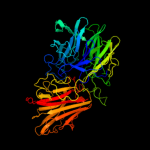
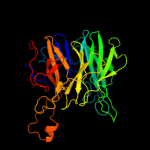
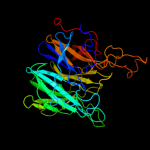
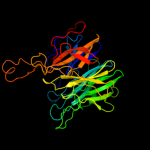
1 c2exiA_
100.0
57
PDB header: hydrolaseChain: A: PDB Molecule: beta-d-xylosidase;PDBTitle: structure of the family43 beta-xylosidase d15g mutant from geobacillus2 stearothermophilus
2 c1yifC_
100.0
54
PDB header: hydrolaseChain: C: PDB Molecule: beta-1,4-xylosidase;PDBTitle: crystal structure of beta-1,4-xylosidase from bacillus2 subtilis, new york structural genomics consortium
3 c1yrzB_
100.0
48
PDB header: hydrolaseChain: B: PDB Molecule: xylan beta-1,4-xylosidase;PDBTitle: crystal structure of xylan beta-1,4-xylosidase from bacillus2 halodurans c-125
4 c1yi7C_
100.0
54
PDB header: hydrolaseChain: C: PDB Molecule: beta-xylosidase, family 43 glycosyl hydrolase;PDBTitle: beta-d-xylosidase (selenomethionine) xynd from clostridium2 acetobutylicum
5 d2exha2
100.0
60
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like6 d1yifa2
100.0
58
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like7 d1y7ba2
100.0
57
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like8 d1yrza2
100.0
54
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like9 c3lv4B_
100.0
23
PDB header: hydrolaseChain: B: PDB Molecule: glycoside hydrolase yxia;PDBTitle: crystal structure of the glycoside hydrolase, family 43 yxia2 protein from bacillus licheniformis. northeast structural3 genomics consortium target bir14.
10 c3akgA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: putative secreted alpha l-arabinofuranosidase ii;PDBTitle: crystal structure of exo-1,5-alpha-l-arabinofuranosidase complexed2 with alpha-1,5-l-arabinofuranobiose
11 c3c7hA_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-xylanase;PDBTitle: crystal structure of glycoside hydrolase family 43 arabinoxylan2 arabinofuranohydrolase from bacillus subtilis in complex with axos-4-3 0.5.
12 c3cpnA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: beta-xylosidase, family 43 glycosyl hydrolase;PDBTitle: crystal structure of beta-xylosidase, family 43 glycosyl2 hydrolase from clostridium acetobutylicum
13 d1gyha_
100.0
22
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like14 c3kstA_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-xylanase;PDBTitle: crystal structure of endo-1,4-beta-xylanase (np_811807.1)2 from bacteroides thetaiotaomicron vpi-5482 at 1.70 a3 resolution
15 d1wl7a1
100.0
22
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like16 d1uv4a1
100.0
21
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like17 c3p2nB_
100.0
18
PDB header: hydrolaseChain: B: PDB Molecule: 3,6-anhydro-alpha-l-galactosidase;PDBTitle: discovery and structural characterization of a new glycoside hydrolase2 family abundant in coastal waters that was annotated as 'hypothetical3 protein'
18 c3qefB_
100.0
23
PDB header: hydrolaseChain: B: PDB Molecule: beta-xylosidase/alpha-l-arabinfuranosidase, gly43n;PDBTitle: the structure and function of an arabinan-specific alpha-1,2-2 arabinofuranosidase identified from screening the activities of3 bacterial gh43 glycoside hydrolases
19 c3qz4B_
100.0
23
PDB header: hydrolaseChain: B: PDB Molecule: endo-1,4-beta-xylanase d;PDBTitle: crystal structure of an endo-1,4-beta-xylanase d (bt_3675) from2 bacteroides thetaiotaomicron vpi-5482 at 1.74 a resolution
20 d1y7ba1
100.0
50
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Beta-D-xylosidase C-terminal domain-like21 d2exha1
not modelled
100.0
54
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Beta-D-xylosidase C-terminal domain-like22 d1yifa1
not modelled
100.0
49
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Beta-D-xylosidase C-terminal domain-like23 d1yrza1
not modelled
100.0
37
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Beta-D-xylosidase C-terminal domain-like24 c3nqhA_
not modelled
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase;PDBTitle: crystal structure of a glycosyl hydrolase (bt_2959) from bacteroides2 thetaiotaomicron vpi-5482 at 2.11 a resolution
25 c3o12A_
not modelled
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yjl217w;PDBTitle: the crystal structure of a functionally unknown protein from2 saccharomyces cerevisiae.
26 c1w2tE_
not modelled
99.9
15
PDB header: hydrolaseChain: E: PDB Molecule: beta fructosidase;PDBTitle: beta-fructosidase from thermotoga maritima in complex with2 raffinose
27 c3mepC_
not modelled
99.9
18
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein eca2234;PDBTitle: crystal structure of eca2234 protein from erwinia2 carotovora, northeast structural genomics consortium target3 ewr44
28 c3pijA_
not modelled
99.9
15
PDB header: hydrolaseChain: A: PDB Molecule: beta-fructofuranosidase;PDBTitle: beta-fructofuranosidase from bifidobacterium longum - complex with2 fructose
29 d1uypa2
not modelled
99.8
16
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: Glycosyl hydrolases family 32 N-terminal domain30 c1y9gA_
not modelled
99.8
15
PDB header: hydrolaseChain: A: PDB Molecule: exo-inulinase;PDBTitle: crystal structure of exo-inulinase from aspergillus awamori complexed2 with fructose
31 c3kf5A_
not modelled
99.8
12
PDB header: hydrolaseChain: A: PDB Molecule: invertase;PDBTitle: structure of invertase from schwanniomyces occidentalis
32 d1vkda_
not modelled
99.8
12
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: TM1225-like predicted glycosylases33 c2aezA_
not modelled
99.7
14
PDB header: hydrolaseChain: A: PDB Molecule: fructan 1-exohydrolase iia;PDBTitle: crystal structure of fructan 1-exohydrolase iia (e201q) from cichorium2 intybus in complex with 1-kestose
34 c3bykA_
not modelled
99.7
17
PDB header: transferaseChain: A: PDB Molecule: levansucrase;PDBTitle: crystal structure of b. subtilis levansucrase mutant d247a
35 c3ugfB_
not modelled
99.7
14
PDB header: transferaseChain: B: PDB Molecule: sucrose:(sucrose/fructan) 6-fructosyltransferase;PDBTitle: crystal structure of a 6-sst/6-sft from pachysandra terminalis
36 c2ac1A_
not modelled
99.7
12
PDB header: hydrolaseChain: A: PDB Molecule: invertase;PDBTitle: crystal structure of a cell-wall invertase from arabidopsis thaliana
37 d1oyga_
not modelled
99.7
18
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: Levansucrase38 c3lemA_
not modelled
99.6
15
PDB header: hydrolaseChain: A: PDB Molecule: fructosyltransferase;PDBTitle: crystal structure of fructosyltransferase (d191a) from a. japonicus in2 complex with nystose
39 c2yfrA_
not modelled
99.6
17
PDB header: transferaseChain: A: PDB Molecule: levansucrase;PDBTitle: crystal structure of inulosucrase from lactobacillus2 johnsonii ncc533
40 d1y4wa2
not modelled
99.5
18
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: Glycosyl hydrolases family 32 N-terminal domain41 c3tawA_
not modelled
99.4
16
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical glycoside hydrolase;PDBTitle: crystal structure of a hypothetical glycoside hydrolase (bdi_3141)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
42 c3qc2A_
not modelled
99.4
17
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase;PDBTitle: crystal structure of a glycosyl hydrolase (bacova_03624) from2 bacteroides ovatus at 2.30 a resolution
43 c1w18A_
not modelled
99.4
16
PDB header: transferaseChain: A: PDB Molecule: levansucrase;PDBTitle: crystal structure of levansucrase from gluconacetobacter2 diazotrophicus
44 c3r67A_
not modelled
99.3
15
PDB header: hydrolaseChain: A: PDB Molecule: putative glycosidase;PDBTitle: crystal structure of a putative glycosidase (bt_4094) from bacteroides2 thetaiotaomicron vpi-5482 at 2.30 a resolution
45 c2xcyA_
not modelled
95.9
15
PDB header: hydrolaseChain: A: PDB Molecule: extracellular sialidase/neuraminidase, putative;PDBTitle: crystal structure of aspergillus fumigatus sialidase
46 c3a72A_
not modelled
95.7
16
PDB header: hydrolaseChain: A: PDB Molecule: exo-arabinanase;PDBTitle: high resolution structure of penicillium chrysogenum alpha-l-2 arabinanase complexed with arabinobiose
47 d2ayha_
not modelled
94.1
15
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 1648 c3i4iA_
not modelled
93.5
11
PDB header: hydrolaseChain: A: PDB Molecule: 1,3-1,4-beta-glucanase;PDBTitle: crystal structure of a prokaryotic beta-1,3-1,4-glucanase (lichenase)2 derived from a mouse hindgut metagenome
49 d1gbga_
not modelled
91.4
12
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 1650 c1upsB_
not modelled
91.0
17
PDB header: glycosyl hydrolaseChain: B: PDB Molecule: glcnac-alpha-1,4-gal-releasing endo-beta-PDBTitle: glcnac[alpha]1-4gal releasing endo-[beta]-galactosidase2 from clostridium perfringens
51 c2w5oA_
not modelled
90.6
13
PDB header: hydrolaseChain: A: PDB Molecule: alpha-l-arabinofuranosidase;PDBTitle: complex structure of the gh93 alpha-l-arabinofuranosidase2 of fusarium graminearum with arabinobiose
52 c3juuA_
not modelled
89.4
18
PDB header: hydrolase/carbohydrateChain: A: PDB Molecule: porphyranase b;PDBTitle: crystal structure of porphyranase b (porb) from zobellia2 galactanivorans
53 c1yo8A_
not modelled
88.2
15
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-2;PDBTitle: structure of the c-terminal domain of human thrombospondin-2
54 c3o5sA_
not modelled
87.8
12
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucanase;PDBTitle: crystal structure of the endo-beta-1,3-1,4 glucanase from bacillus2 subtilis (strain 168)
55 c3fbyC_
not modelled
85.1
15
PDB header: cell adhesionChain: C: PDB Molecule: cartilage oligomeric matrix protein;PDBTitle: the crystal structure of the signature domain of cartilage oligomeric2 matrix protein.
56 c3rq0A_
not modelled
83.6
12
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolases family protein 16;PDBTitle: the crystal structure of a glycosyl hydrolases (gh) family protein 162 from mycobacterium smegmatis str. mc2 155
57 d1o4ya_
not modelled
82.9
14
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 1658 c2w38A_
not modelled
77.1
15
PDB header: transferaseChain: A: PDB Molecule: sialidase;PDBTitle: crystal structure of the pseudaminidase from pseudomonas2 aeruginosa
59 d1w8oa3
not modelled
76.4
15
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)60 c3zr6A_
not modelled
75.8
16
PDB header: hydrolaseChain: A: PDB Molecule: galactocerebrosidase;PDBTitle: structure of galactocerebrosidase from mouse in complex with2 galactose
61 c3dgtA_
not modelled
72.1
23
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,3-beta-glucanase;PDBTitle: the 1.5 a crystal structure of endo-1,3-beta-glucanase from2 streptomyces sioyaensis
62 d1upsa1
not modelled
71.7
18
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 1663 d1o4za_
not modelled
67.2
9
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 1664 c1o4zA_
not modelled
67.2
9
PDB header: hydrolaseChain: A: PDB Molecule: beta-agarase b;PDBTitle: the three-dimensional structure of beta-agarase b from2 zobellia galactanivorans
65 c2berA_
not modelled
66.8
16
PDB header: hydrolaseChain: A: PDB Molecule: bacterial sialidase;PDBTitle: y370g active site mutant of the sialidase from2 micromonospora viridifaciens in complex with beta-neu5ac3 (sialic acid).
66 d2b4wa1
not modelled
64.6
10
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: LmjF10.1260-like67 d1y4wa1
not modelled
63.1
12
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 32 C-terminal domain68 d2ah2a2
not modelled
62.8
15
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)69 c2w20B_
not modelled
59.9
13
PDB header: hydrolaseChain: B: PDB Molecule: sialidase a;PDBTitle: structure of the catalytic domain of the native nana2 sialidase from streptococcus pneumoniae
70 c1ux6A_
not modelled
55.9
14
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-1;PDBTitle: structure of a thrombospondin c-terminal fragment reveals a2 novel calcium core in the type 3 repeats
71 c3h3lB_
not modelled
55.0
14
PDB header: hydrolaseChain: B: PDB Molecule: putative sugar hydrolase;PDBTitle: crystal structure of putative sugar hydrolase (yp_001304206.1) from2 parabacteroides distasonis atcc 8503 at 1.59 a resolution
72 c3azyA_
not modelled
54.7
18
PDB header: hydrolaseChain: A: PDB Molecule: laminarinase;PDBTitle: crystal structure of the laminarinase catalytic domain from thermotoga2 maritima msb8
73 d1fbla1
not modelled
50.1
16
Fold: 4-bladed beta-propellerSuperfamily: Hemopexin-like domainFamily: Hemopexin-like domain74 c2vk7A_
not modelled
44.1
11
PDB header: hydrolaseChain: A: PDB Molecule: exo-alpha-sialidase;PDBTitle: the structure of clostridium perfringens nani sialidase and2 its catalytic intermediates
75 c3nmbA_
not modelled
44.0
12
PDB header: hydrolaseChain: A: PDB Molecule: putative sugar hydrolase;PDBTitle: crystal structure of a putative sugar hydrolase (bacova_03189) from2 bacteroides ovatus at 2.40 a resolution
76 c3ecqA_
not modelled
42.0
11
PDB header: hydrolaseChain: A: PDB Molecule: endo-alpha-n-acetylgalactosaminidase;PDBTitle: endo-alpha-n-acetylgalactosaminidase from streptococcus pneumoniae:2 semet structure
77 d1dypa_
not modelled
39.0
13
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 1678 d1n1ta2
not modelled
36.0
15
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)79 c3u1xA_
not modelled
35.6
11
PDB header: hydrolaseChain: A: PDB Molecule: putative glycosyl hydrolase;PDBTitle: crystal structure of a putative glycosyl hydrolase (bdi_1869) from2 parabacteroides distasonis atcc 8503 at 1.70 a resolution
80 c1su3A_
not modelled
35.4
16
PDB header: hydrolaseChain: A: PDB Molecule: interstitial collagenase;PDBTitle: x-ray structure of human prommp-1: new insights into2 collagenase action
81 d1w0pa3
not modelled
35.3
9
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)82 c2vvzA_
not modelled
34.6
13
PDB header: hydrolaseChain: A: PDB Molecule: sialidase a;PDBTitle: structure of the catalytic domain of streptococcus2 pneumoniae sialidase nana
83 d3sila_
not modelled
30.5
15
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)84 d1ux6a1
not modelled
29.1
17
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Thrombospondin C-terminal domain85 d1pexa_
not modelled
28.1
11
Fold: 4-bladed beta-propellerSuperfamily: Hemopexin-like domainFamily: Hemopexin-like domain86 c3oyoB_
not modelled
23.8
11
PDB header: plant proteinChain: B: PDB Molecule: hemopexin fold protein cp4;PDBTitle: crystal structure of hemopexin fold protein cp4 from cow pea
87 d2a6za1
not modelled
23.5
12
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Lectin leg-like88 d1gena_
not modelled
23.1
9
Fold: 4-bladed beta-propellerSuperfamily: Hemopexin-like domainFamily: Hemopexin-like domain89 d1su3a2
not modelled
20.6
14
Fold: 4-bladed beta-propellerSuperfamily: Hemopexin-like domainFamily: Hemopexin-like domain90 c1gxdA_
not modelled
20.5
10
PDB header: hydrolaseChain: A: PDB Molecule: 72 kda type iv collagenase;PDBTitle: prommp-2/timp-2 complex
91 c3ilfA_
not modelled
19.2
13
PDB header: hydrolase/carbohydrateChain: A: PDB Molecule: porphyranase a;PDBTitle: crystal structure of porphyranase a (pora) in complex with2 neo-porphyrotetraose
92 c3c7xA_
not modelled
17.6
17
PDB header: hydrolaseChain: A: PDB Molecule: matrix metalloproteinase-14;PDBTitle: hemopexin-like domain of matrix metalloproteinase 14
93 c3hbkA_
not modelled
17.2
14
PDB header: hydrolaseChain: A: PDB Molecule: putative glycosyl hydrolase;PDBTitle: crystal structure of putative glycosyl hydrolase, was domain of2 unknown function (duf1080) (yp_001302580.1) from parabacteroides3 distasonis atcc 8503 at 2.36 a resolution
94 c3eypB_
not modelled
16.4
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative alpha-l-fucosidase;PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
95 c3lp9C_
not modelled
16.2
15
PDB header: plant proteinChain: C: PDB Molecule: ls-24;PDBTitle: crystal structure of ls24, a seed albumin from lathyrus2 sativus
96 c2v5dA_
not modelled
16.2
25
PDB header: hydrolaseChain: A: PDB Molecule: o-glcnacase nagj;PDBTitle: structure of a family 84 glycoside hydrolase and a family2 32 carbohydrate-binding module in tandem from clostridium3 perfringens.
97 d1czsa_
not modelled
14.5
31
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Discoidin domain (FA58C, coagulation factor 5/8 C-terminal domain)98 c2xqxB_
not modelled
13.8
13
PDB header: sugar binding proteinChain: B: PDB Molecule: endo-beta-n-acetylglucosaminidase d;PDBTitle: structure of the family 32 carbohydrate-binding module from2 streptococcus pneumoniae endod
99 c3jtoE_
not modelled
13.7
33
PDB header: protein bindingChain: E: PDB Molecule: adapter protein meca 2;PDBTitle: crystal structure of the c-terminal domain of ypbh