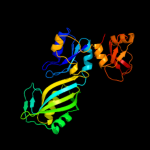
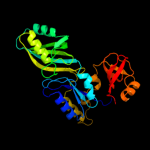
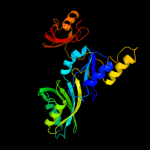
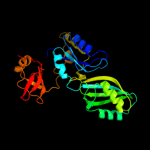
1 c1k8wA_
100.0
100
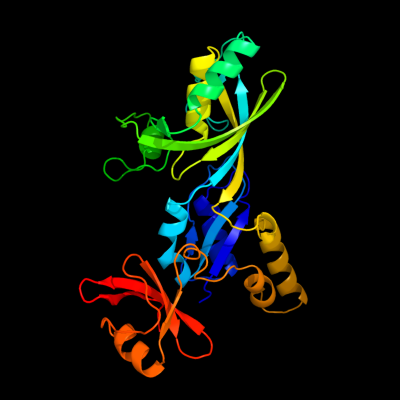
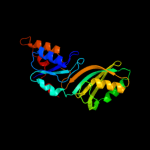
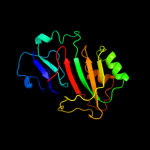
PDB header: lyase/rnaChain: A: PDB Molecule: trna pseudouridine synthase b;PDBTitle: crystal structure of the e. coli pseudouridine synthase2 trub bound to a t stem-loop rna
2 d1k8wa5
100.0
100
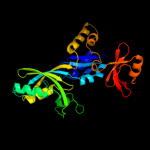
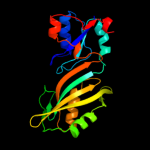
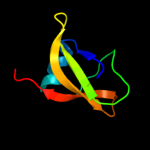
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB3 d1r3ea2
100.0
33
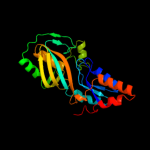
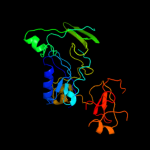
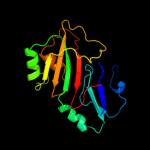
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB4 c1sgvA_
100.0
32
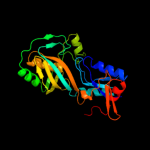
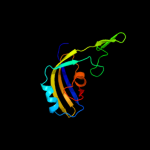
PDB header: lyaseChain: A: PDB Molecule: trna pseudouridine synthase b;PDBTitle: structure of trna psi55 pseudouridine synthase (trub)
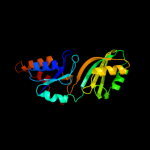
5 c2ey4A_
100.0
29
PDB header: isomerase/biosynthetic proteinChain: A: PDB Molecule: probable trna pseudouridine synthase b;PDBTitle: crystal structure of a cbf5-nop10-gar1 complex
6 c3uaiA_
100.0
28
PDB header: isomerase/chaperoneChain: A: PDB Molecule: h/aca ribonucleoprotein complex subunit 4;PDBTitle: structure of the shq1-cbf5-nop10-gar1 complex from saccharomyces2 cerevisiae
7 c2apoA_
100.0
31
PDB header: isomerase/rna binding proteinChain: A: PDB Molecule: probable trna pseudouridine synthase b;PDBTitle: crystal structure of the methanococcus jannaschii cbf52 nop10 complex
8 d2ey4a2
100.0
32
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB9 d1sgva2
100.0
37
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB10 c1ze2B_
100.0
29
PDB header: lyase/rnaChain: B: PDB Molecule: trna pseudouridine synthase b;PDBTitle: conformational change of pseudouridine 55 synthase upon its2 association with rna substrate
11 d2apoa2
100.0
32
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB12 c2omlA_
99.2
20
PDB header: isomeraseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase e;PDBTitle: crystal structure of e. coli pseudouridine synthase rlue
13 d1k8wa3
99.1
100
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain14 c2olwB_
98.9
20
PDB header: isomeraseChain: B: PDB Molecule: ribosomal large subunit pseudouridine synthase e;PDBTitle: crystal structure of e. coli pseudouridine synthase rlue
15 c2v9kA_
98.3
17
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein flj32312;PDBTitle: crystal structure of human pus10, a novel pseudouridine2 synthase.
16 d1vioa1
98.2
23
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD17 c2gmlA_
98.1
22
PDB header: isomeraseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase f;PDBTitle: crystal structure of catalytic domain of e.coli rluf
18 c3dh3C_
98.1
18
PDB header: isomerase/rnaChain: C: PDB Molecule: ribosomal large subunit pseudouridine synthase f;PDBTitle: crystal structure of rluf in complex with a 22 nucleotide2 rna substrate
19 d1kska4
98.0
19
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD20 c1vioA_
97.7
24
PDB header: lyaseChain: A: PDB Molecule: ribosomal small subunit pseudouridine synthase a;PDBTitle: crystal structure of pseudouridylate synthase
21 d1v9ka_
not modelled
97.5
20
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD22 c3zv0D_
not modelled
97.5
19
PDB header: cell cycleChain: D: PDB Molecule: h/aca ribonucleoprotein complex subunit 4;PDBTitle: structure of the shq1p-cbf5p complex
23 c2i82D_
not modelled
97.4
17
PDB header: lyase/rnaChain: D: PDB Molecule: ribosomal large subunit pseudouridine synthase a;PDBTitle: crystal structure of pseudouridine synthase rlua: indirect2 sequence readout through protein-induced rna structure
24 c1kskA_
not modelled
97.3
21
PDB header: lyaseChain: A: PDB Molecule: ribosomal small subunit pseudouridine synthase a;PDBTitle: structure of rsua
25 c1qyuA_
not modelled
97.3
24
PDB header: lyaseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase d;PDBTitle: structure of the catalytic domain of 23s rrna pseudouridine2 synthase rlud
26 d1v9fa_
not modelled
97.3
23
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD27 c1v9fA_
not modelled
97.3
23
PDB header: lyaseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase d;PDBTitle: crystal structure of catalytic domain of pseudouridine2 synthase rlud from escherichia coli
28 d2apoa1
not modelled
95.5
23
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain29 d1r3ea1
not modelled
93.5
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain30 d2ey4a1
not modelled
93.1
13
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain31 d1sgva1
not modelled
90.2
18
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain32 c1q7hA_
not modelled
75.1
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: structure of a conserved pua domain protein from thermoplasma2 acidophilum
33 c3d79A_
not modelled
74.1
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein ph0734;PDBTitle: crystal structure of hypothetical protein ph0734.1 from2 hyperthermophilic archaea pyrococcus horikoshii ot3
34 d1iq8a3
not modelled
73.3
16
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain35 d2as0a1
not modelled
68.7
21
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Hypothetical RNA methyltransferase domain (HRMD)36 c1sb7A_
not modelled
67.7
12
PDB header: lyaseChain: A: PDB Molecule: trna pseudouridine synthase d;PDBTitle: crystal structure of the e.coli pseudouridine synthase trud
37 d1szwa_
not modelled
67.5
10
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: tRNA pseudouridine synthase TruD38 d2b78a1
not modelled
59.6
12
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Hypothetical RNA methyltransferase domain (HRMD)39 c2b78A_
not modelled
57.3
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein smu.776;PDBTitle: a putative sam-dependent methyltransferase from2 streptococcus mutans
40 d1q7ha1
not modelled
54.4
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain41 c3r90E_
not modelled
50.6
11
PDB header: rna binding proteinChain: E: PDB Molecule: malignant t cell-amplified sequence 1;PDBTitle: crystal structure of malignant t cell-amplified sequence 1 protein
42 c1zs7A_
not modelled
44.3
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ape0525;PDBTitle: the structure of gene product ape0525 from aeropyrum pernix
43 c2gagC_
not modelled
37.2
15
PDB header: oxidoreductaseChain: C: PDB Molecule: heterotetrameric sarcosine oxidase gamma-subunit;PDBTitle: heteroteterameric sarcosine: structure of a diflavin metaloenzyme at2 1.85 a resolution
44 c2frxD_
not modelled
35.0
21
PDB header: transferaseChain: D: PDB Molecule: hypothetical protein yebu;PDBTitle: crystal structure of yebu, a m5c rna methyltransferase from e.coli
45 c3m4xA_
not modelled
34.6
18
PDB header: transferaseChain: A: PDB Molecule: nol1/nop2/sun family protein;PDBTitle: structure of a ribosomal methyltransferase
46 c2xzmW_
not modelled
34.4
23
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s4;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
47 c2hjgA_
not modelled
29.6
8
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein enga;PDBTitle: the crystal structure of the b. subtilis yphc gtpase in2 complex with gdp
48 d2a2pa1
not modelled
28.4
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Selenoprotein W-related49 d2jn9a1
not modelled
28.2
9
Fold: Split barrel-likeSuperfamily: YkvR-likeFamily: YkvR-like50 d1o7qa_
not modelled
24.8
21
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: alpha-1,3-galactosyltransferase-like51 d2cx1a1
not modelled
22.1
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain52 d1lzia_
not modelled
21.7
15
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: alpha-1,3-galactosyltransferase-like53 d2gx8a1
not modelled
21.5
17
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like54 c1z2zB_
not modelled
18.9
16
PDB header: lyaseChain: B: PDB Molecule: probable trna pseudouridine synthase d;PDBTitle: crystal structure of the putative trna pseudouridine2 synthase d (trud) from methanosarcina mazei, northeast3 structural genomics target mar1
55 d1mkya3
not modelled
18.9
3
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Probable GTPase Der, C-terminal domainFamily: Probable GTPase Der, C-terminal domain56 c2gx8B_
not modelled
18.3
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal stucture of bacillus cereus protein related to nif3
57 d1k7ia2
not modelled
16.6
33
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Serralysin-like metalloprotease, catalytic (N-terminal) domain58 c3czxA_
not modelled
15.3
50
PDB header: hydrolaseChain: A: PDB Molecule: putative n-acetylmuramoyl-l-alanine amidase;PDBTitle: the crystal structure of the putative n-acetylmuramoyl-l-2 alanine amidase from neisseria meningitidis
59 d1g9ka2
not modelled
14.5
39
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Serralysin-like metalloprotease, catalytic (N-terminal) domain60 c1mkyA_
not modelled
14.3
3
PDB header: ligand binding proteinChain: A: PDB Molecule: probable gtp-binding protein enga;PDBTitle: structural analysis of the domain interactions in der, a2 switch protein containing two gtpase domains
61 c3m6wA_
not modelled
13.7
22
PDB header: transferaseChain: A: PDB Molecule: rrna methylase;PDBTitle: multi-site-specific 16s rrna methyltransferase rsmf from thermus2 thermophilus in space group p21212 in complex with s-adenosyl-l-3 methionine
62 c3or5A_
not modelled
13.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein, thioredoxin familyPDBTitle: crystal structure of thiol:disulfide interchange protein, thioredoxin2 family protein from chlorobium tepidum tls
63 c3eykA_
not modelled
13.2
15
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin ha1 chain;PDBTitle: structure of influenza haemagglutinin in complex with an2 inhibitor of membrane fusion
64 c3cmiA_
not modelled
11.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxiredoxin hyr1;PDBTitle: crystal structure of glutathione-dependent phospholipid peroxidase2 hyr1 from the yeast saccharomyces cerevisiae
65 c1iq8B_
not modelled
11.5
16
PDB header: transferaseChain: B: PDB Molecule: archaeosine trna-guanine transglycosylase;PDBTitle: crystal structure of archaeosine trna-guanine2 transglycosylase from pyrococcus horikoshii
66 c2nydB_
not modelled
11.5
13
PDB header: unknown functionChain: B: PDB Molecule: upf0135 protein sa1388;PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
67 d1sata2
not modelled
11.3
33
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Serralysin-like metalloprotease, catalytic (N-terminal) domain68 d3saka_
not modelled
11.1
29
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain69 c2owlA_
not modelled
10.0
12
PDB header: recombinationChain: A: PDB Molecule: recombination-associated protein rdgc;PDBTitle: crystal structure of e. coli rdgc
70 c3ne8A_
not modelled
10.0
50
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase;PDBTitle: the crystal structure of a domain from n-acetylmuramoyl-l-alanine2 amidase of bartonella henselae str. houston-1
71 d1muga_
not modelled
9.1
19
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Mug-like72 c3kbgA_
not modelled
9.0
15
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s4e;PDBTitle: crystal structure of the 30s ribosomal protein s4e from2 thermoplasma acidophilum. northeast structural genomics3 consortium target tar28.
73 c2d07A_
not modelled
8.8
16
PDB header: hydrolaseChain: A: PDB Molecule: g/t mismatch-specific thymine dna glycosylase;PDBTitle: crystal structure of sumo-3-modified thymine-dna glycosylase
74 d1jwqa_
not modelled
8.3
50
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: N-acetylmuramoyl-L-alanine amidase-like75 d2gy9i1
not modelled
8.3
18
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components76 c1yp3C_
not modelled
7.7
24
PDB header: transferaseChain: C: PDB Molecule: glucose-1-phosphate adenylyltransferase smallPDBTitle: crystal structure of potato tuber adp-glucose2 pyrophosphorylase in complex with atp
77 c2xs4A_
not modelled
7.4
28
PDB header: hydrolaseChain: A: PDB Molecule: karilysin protease;PDBTitle: structure of karilysin catalytic mmp domain in complex with2 magnesium
78 d1xova2
not modelled
7.3
29
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: N-acetylmuramoyl-L-alanine amidase-like79 c1pk1B_
not modelled
7.3
12
PDB header: transcription repressionChain: B: PDB Molecule: sex comb on midleg cg9495-pa;PDBTitle: hetero sam domain structure of ph and scm.
80 d1hv5a_
not modelled
7.3
26
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain81 c1mszA_
not modelled
7.3
21
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein smubp-2;PDBTitle: solution structure of the r3h domain from human smubp-2
82 d1msza_
not modelled
7.3
21
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain83 c2vn1A_
not modelled
7.1
13
PDB header: isomeraseChain: A: PDB Molecule: 70 kda peptidylprolyl isomerase;PDBTitle: crystal structure of the fk506-binding domain of plasmodium2 falciparum fkbp35 in complex with fk506
84 d1cgla_
not modelled
7.0
22
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain85 c3lfmA_
not modelled
7.0
46
PDB header: oxidoreductaseChain: A: PDB Molecule: protein fto;PDBTitle: crystal structure of the fat mass and obesity associated (fto) protein2 reveals basis for its substrate specificity
86 d1bqqm_
not modelled
7.0
32
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain87 d1g0da2
not modelled
6.8
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Transglutaminase, two C-terminal domainsFamily: Transglutaminase, two C-terminal domains88 d1nmpa_
not modelled
6.7
6
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like89 d2fg9a1
not modelled
6.6
19
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like90 d1s1qa_
not modelled
6.6
18
Fold: UBC-likeSuperfamily: UBC-likeFamily: UEV domain91 c3eytA_
not modelled
6.6
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein spoa0173;PDBTitle: crystal structure of thioredoxin-like superfamily protein spoa0173
92 c2e1cA_
not modelled
6.4
16
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
93 d2ovxa1
not modelled
6.2
22
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain94 d2b5xa1
not modelled
6.2
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like95 c2obiA_
not modelled
6.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: phospholipid hydroperoxide glutathionePDBTitle: crystal structure of the selenocysteine to cysteine mutant2 of human phospholipid hydroperoxide glutathione peroxidase3 (gpx4)
96 d1nkva_
not modelled
6.0
8
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Hypothetical Protein YjhP97 d1r9ha_
not modelled
5.8
16
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase98 c2l4bA_
not modelled
5.7
24
PDB header: transferaseChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of a putative acyl carrier protein from anaplasma2 phagocytophilum. seattle structural genomics center for infectious3 disease target anpha.01018.a
99 c3iz6D_
not modelled
5.7
14
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein s4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome