1 c1rs0A_
99.6
15
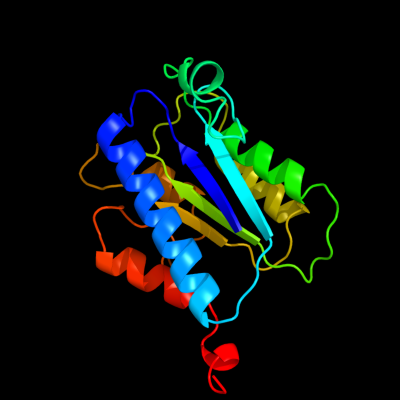
PDB header: hydrolaseChain: A: PDB Molecule: complement factor b;PDBTitle: crystal structure analysis of the bb segment of factor b2 complexed with di-isopropyl-phosphate (dip)
2 c2ok5A_
99.5
17
PDB header: hydrolaseChain: A: PDB Molecule: complement factor b;PDBTitle: human complement factor b
3 d2ok5a1
99.5
15
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain4 c2i6sA_
99.4
18
PDB header: hydrolaseChain: A: PDB Molecule: complement c2a fragment;PDBTitle: complement component c2a
5 c2x5nA_
99.4
11
PDB header: nuclear proteinChain: A: PDB Molecule: 26s proteasome regulatory subunit rpn10;PDBTitle: crystal structure of the sprpn10 vwa domain
6 c2iueA_
99.4
13
PDB header: membrane proteinChain: A: PDB Molecule: pactolus i-domain;PDBTitle: pactolus i-domain: functional switching of the rossmann2 fold
7 d1jeyb2
99.3
13
Fold: vWA-likeSuperfamily: vWA-likeFamily: Ku80 subunit N-terminal domain8 d1ijba_
99.2
11
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain9 c3ibsA_
99.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein batb;PDBTitle: crystal structure of conserved hypothetical protein batb from2 bacteroides thetaiotaomicron
10 c2nvoA_
99.1
16
PDB header: rna binding proteinChain: A: PDB Molecule: ro sixty-related protein, rsr;PDBTitle: crystal structure of deinococcus radiodurans ro (rsr) protein
11 d1jeya2
99.1
12
Fold: vWA-likeSuperfamily: vWA-likeFamily: Ku70 subunit N-terminal domain12 d1yvra2
99.1
14
Fold: vWA-likeSuperfamily: vWA-likeFamily: RoRNP C-terminal domain-like13 d1u0oc1
99.1
11
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain14 c2x31F_
99.1
21
PDB header: ligaseChain: F: PDB Molecule: magnesium-chelatase 60 kda subunit;PDBTitle: modelling of the complex between subunits bchi and bchd of magnesium2 chelatase based on single-particle cryo-em reconstruction at 7.5 ang
15 d1shux_
99.0
14
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain16 d1q0pa_
99.0
16
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain17 c2xggB_
99.0
16
PDB header: hydrolaseChain: B: PDB Molecule: microneme protein 2;PDBTitle: structure of toxoplasma gondii micronemal protein 2 a_i2 domain
18 c3n2nC_
98.9
14
PDB header: toxin receptorChain: C: PDB Molecule: anthrax toxin receptor 1;PDBTitle: the crystal structure of tumor endothelial marker 8 (tem8)2 extracellular domain
19 c1jeyB_
98.9
13
PDB header: dna binding protein/dnaChain: B: PDB Molecule: ku80;PDBTitle: crystal structure of the ku heterodimer bound to dna
20 c2b2xB_
98.9
12
PDB header: immune systemChain: B: PDB Molecule: integrin alpha-1;PDBTitle: vla1 rdeltah i-domain complexed with a quadruple mutant of the aqc22 fab
21 d1v7pc_
not modelled
98.9
11
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain22 d1atza_
not modelled
98.8
13
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain23 d1n3ya_
not modelled
98.8
15
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain24 c3gxbB_
not modelled
98.8
10
PDB header: cell adhesionChain: B: PDB Molecule: von willebrand factor;PDBTitle: crystal structure of vwf a2 domain
25 d1pt6a_
not modelled
98.8
11
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain26 d1mjna_
not modelled
98.8
15
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain27 d1ck4a_
not modelled
98.7
11
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain28 d1mf7a_
not modelled
98.7
12
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain29 d1pd0a3
not modelled
98.6
13
Fold: vWA-likeSuperfamily: vWA-likeFamily: Trunk domain of Sec23/2430 c3ragA_
not modelled
98.4
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein aaci_0196 from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
31 d1tyeb2
not modelled
98.3
16
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain32 c1yvrA_
98.2
15
PDB header: rna binding proteinChain: A: PDB Molecule: 60-kda ss-a/ro ribonucleoprotein;PDBTitle: ro autoantigen
33 c3v4pB_
not modelled
98.0
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-7;PDBTitle: crystal structure of a4b7 headpiece complexed with fab act-1
34 c3fcuB_
not modelled
98.0
16
PDB header: cell adhesion/blood clottingChain: B: PDB Molecule: integrin beta-3;PDBTitle: structure of headpiece of integrin aiibb3 in open conformation
35 c1jeqA_
not modelled
97.5
13
PDB header: dna binding proteinChain: A: PDB Molecule: ku70;PDBTitle: crystal structure of the ku heterodimer
36 c1u8cB_
not modelled
97.2
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
37 c3k6sB_
not modelled
96.1
14
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-2;PDBTitle: structure of integrin alphaxbeta2 ectodomain
38 c1pd0A_
not modelled
96.0
13
PDB header: transport proteinChain: A: PDB Molecule: protein transport protein sec24;PDBTitle: crystal structure of the copii coat subunit, sec24,2 complexed with a peptide from the snare protein sed53 (yeast syntaxin-5)
39 c1m2vB_
not modelled
95.2
14
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24;PDBTitle: crystal structure of the yeast sec23/24 heterodimer
40 c3eg9B_
not modelled
89.8
14
PDB header: protein transportChain: B: PDB Molecule: sec24 related gene family, member d;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23/24 bound to the transport signal sequence of membrin
41 c3egxB_
not modelled
89.1
13
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24a;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23a/24a complexed with the snare protein sec22b and3 bound to the transport signal sequence of the snare protein4 bet1
42 c3eg9A_
not modelled
84.2
15
PDB header: protein transportChain: A: PDB Molecule: protein transport protein sec23a;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23/24 bound to the transport signal sequence of membrin
43 d2qtva3
not modelled
82.5
15
Fold: vWA-likeSuperfamily: vWA-likeFamily: Trunk domain of Sec23/2444 c3eh2B_
not modelled
78.2
13
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24c;PDBTitle: crystal structure of the human copii-coat protein sec24c
45 c1m2oA_
not modelled
54.8
17
PDB header: protein transport/signaling proteinChain: A: PDB Molecule: protein transport protein sec23;PDBTitle: crystal structure of the sec23-sar1 complex
46 d1h7na_
not modelled
37.0
18
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)47 c3atyA_
not modelled
33.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: prostaglandin f2a synthase;PDBTitle: crystal structure of tcoye
48 c2ftpA_
not modelled
31.8
16
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
49 d1vjia_
not modelled
26.4
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases50 d2c1ha1
not modelled
25.4
10
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)51 d1ja1a2
not modelled
25.2
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like52 d1boba_
not modelled
23.0
8
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT53 c3sggA_
not modelled
21.8
29
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical hydrolase;PDBTitle: crystal structure of a hypothetical hydrolase (bt_2193) from2 bacteroides thetaiotaomicron vpi-5482 at 1.25 a resolution
54 d1icpa_
not modelled
20.7
26
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases55 d1gwja_
not modelled
20.4
23
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases56 c3czpA_
not modelled
20.0
19
PDB header: transferaseChain: A: PDB Molecule: putative polyphosphate kinase 2;PDBTitle: crystal structure of putative polyphosphate kinase 2 from pseudomonas2 aeruginosa pa01
57 c1ydnA_
not modelled
18.3
15
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
58 d1irud_
not modelled
17.5
21
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits59 c1ydoC_
not modelled
15.4
16
PDB header: lyaseChain: C: PDB Molecule: hmg-coa lyase;PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
60 c3komB_
not modelled
14.5
13
PDB header: transferaseChain: B: PDB Molecule: transketolase;PDBTitle: crystal structure of apo transketolase from francisella tularensis
61 d1l6sa_
not modelled
13.4
9
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)62 c3l5aA_
not modelled
13.3
20
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh/flavin oxidoreductase/nadh oxidase;PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
63 d1uiza_
not modelled
12.0
11
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related64 d1w5fa2
not modelled
11.9
11
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain65 c3mwbA_
not modelled
11.7
18
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
66 c3fwtA_
not modelled
11.5
11
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: crystal structure of leishmania major mif2
67 d1gzga_
not modelled
10.8
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)68 c2p0wB_
not modelled
10.8
13
PDB header: transferaseChain: B: PDB Molecule: histone acetyltransferase type b catalytic subunit;PDBTitle: human histone acetyltransferase 1 (hat1)
69 d1vh4a_
not modelled
10.6
25
Fold: Single-stranded right-handed beta-helixSuperfamily: Stabilizer of iron transporter SufDFamily: Stabilizer of iron transporter SufD70 d2vapa2
not modelled
9.9
4
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain71 c2l42A_
not modelled
9.4
19
PDB header: protein bindingChain: A: PDB Molecule: dna-binding protein rap1;PDBTitle: the solution structure of rap1 brct domain from saccharomyces2 cerevisiae
72 c3vcbB_
not modelled
9.2
20
PDB header: viral proteinChain: B: PDB Molecule: rna-directed rna polymerase;PDBTitle: c425s mutant of the c-terminal cytoplasmic domain of non-structural2 protein 4 from mouse hepatitis virus a59
73 d2fcja1
not modelled
9.1
24
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain74 d1q45a_
not modelled
8.8
25
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases75 d1bmlc1
not modelled
8.6
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase76 d1g8fa3
not modelled
8.4
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain77 c3luyA_
not modelled
8.2
7
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
78 d1xhoa_
not modelled
8.1
45
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase79 c2xczA_
not modelled
8.0
12
PDB header: immune systemChain: A: PDB Molecule: possible atls1-like light-inducible protein;PDBTitle: crystal structure of macrophage migration inhibitory factor2 homologue from prochlorococcus marinus
80 c1xhoB_
not modelled
7.7
42
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: chorismate mutase;PDBTitle: chorismate mutase from clostridium thermocellum cth-682
81 c3lukB_
not modelled
7.4
9
PDB header: rna binding proteinChain: B: PDB Molecule: protein argonaute-2;PDBTitle: crystal structure of mid domain from hago2
82 d1rhya2
not modelled
7.4
20
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase83 d1xg8a_
not modelled
7.4
35
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: YuzD-like84 c3gzfD_
not modelled
7.2
28
PDB header: viral proteinChain: D: PDB Molecule: replicase polyprotein 1ab;PDBTitle: structure of the c-terminal domain of nsp4 from feline coronavirus
85 c3dmaA_
not modelled
7.1
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: exopolyphosphatase-related protein;PDBTitle: crystal structure of an exopolyphosphatase-related protein2 from bacteroides fragilis. northeast structural genomics3 target bfr192
86 c2k19A_
not modelled
7.1
15
PDB header: antimicrobial proteinChain: A: PDB Molecule: putative piscicolin 126 immunity protein;PDBTitle: nmr solution structure of pisi
87 c2qtxL_
not modelled
7.0
86
PDB header: rna binding proteinChain: L: PDB Molecule: uncharacterized protein mj1435;PDBTitle: crystal structure of an hfq-like protein from methanococcus2 jannaschii
88 c3b64A_
not modelled
7.0
9
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: macrophage migration inhibitory factor (mif) from2 /leishmania major
89 d2qmwa2
not modelled
7.0
12
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain90 d1nxha_
not modelled
6.8
24
Fold: Hypothetical protein MTH393Superfamily: Hypothetical protein MTH393Family: Hypothetical protein MTH39391 d2cp8a1
not modelled
6.6
44
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain92 c3cs5B_
not modelled
6.5
28
PDB header: photosynthesisChain: B: PDB Molecule: phycobilisome degradation protein nbla;PDBTitle: nbla protein from synechococcus elongatus pcc 7942
93 d2dfaa1
not modelled
6.5
11
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like94 c2h90A_
not modelled
6.5
26
PDB header: oxidoreductaseChain: A: PDB Molecule: xenobiotic reductase a;PDBTitle: xenobiotic reductase a in complex with coumarin
95 c3eukC_
not modelled
6.3
18
PDB header: cell cycleChain: C: PDB Molecule: chromosome partition protein mukb, linker;PDBTitle: crystal structure of muke-mukf(residues 292-443)-mukb(head2 domain)-atpgammas complex, asymmetric dimer
96 d2ae8a2
not modelled
6.3
27
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase97 d1vyra_
not modelled
6.3
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases98 d1pv8a_
not modelled
6.2
20
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)99 d1l4zb_
not modelled
6.2
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase