| 1 | c2gqfA_ |
|
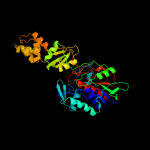 |
100.0 |
56 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hi0933;
PDBTitle: crystal structure of flavoprotein hi0933 from haemophilus influenzae2 rd
|
| 2 | c3v76A_ |
|
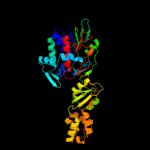 |
100.0 |
52 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavoprotein;
PDBTitle: the crystal structure of a flavoprotein from sinorhizobium meliloti
|
| 3 | c2i0zA_ |
|
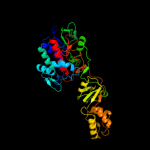 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad(fad)-utilizing dehydrogenases;
PDBTitle: crystal structure of a fad binding protein from bacillus2 cereus, a putative nad(fad)-utilizing dehydrogenases
|
| 4 | c1qo8A_ |
|
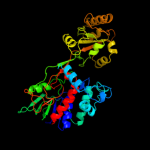 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavocytochrome c3 fumarate reductase;
PDBTitle: the structure of the open conformation of a flavocytochrome2 c3 fumarate reductase
|
| 5 | c1jrxA_ |
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavocytochrome c;
PDBTitle: crystal structure of arg402ala mutant flavocytochrome c32 from shewanella frigidimarina
|
| 6 | c1yq4A_ |
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:succinate dehydrogenase flavoprotein subunit;
PDBTitle: avian respiratory complex ii with 3-nitropropionate and ubiquinone
|
| 7 | c1d4cB_ |
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:flavocytochrome c fumarate reductase;
PDBTitle: crystal structure of the uncomplexed form of the2 flavocytochrome c fumarate reductase of shewanella3 putrefaciens strain mr-1
|
| 8 | d2gqfa1 |
|
 |
100.0 |
66 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
| 9 | c1kf6A_ |
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase flavoprotein;
PDBTitle: e. coli quinol-fumarate reductase with bound inhibitor hqno
|
| 10 | c3p4rM_ |
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:fumarate reductase flavoprotein subunit;
PDBTitle: crystal structure of menaquinol:fumarate oxidoreductase in complex2 with glutarate
|
| 11 | c2aczA_ |
|
 |
100.0 |
21 |
PDB header:oxidoreductase/electron transport
Chain: A: PDB Molecule:succinate dehydrogenase flavoprotein subunit;
PDBTitle: complex ii (succinate dehydrogenase) from e. coli with atpenin a52 inhibitor co-crystallized at the ubiquinone binding site
|
| 12 | c2bs3A_ |
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinol-fumarate reductase flavoprotein subunit a;
PDBTitle: glu c180 -> gln variant quinol:fumarate reductase from2 wolinella succinogenes
|
| 13 | c2fjaC_ |
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:adenylylsulfate reductase, subunit a;
PDBTitle: adenosine 5'-phosphosulfate reductase in complex with2 substrate
|
| 14 | c2e5vA_ |
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-aspartate oxidase;
PDBTitle: crystal structure of l-aspartate oxidase from2 hyperthermophilic archaeon sulfolobus tokodaii
|
| 15 | c1chuA_ |
|
 |
100.0 |
18 |
PDB header:flavoenzyme
Chain: A: PDB Molecule:protein (l-aspartate oxidase);
PDBTitle: structure of l-aspartate oxidase: implications for the2 succinate dehydrogenase/ fumarate reducatse family
|
| 16 | c3gyxA_ |
|
 |
100.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:adenylylsulfate reductase;
PDBTitle: crystal structure of adenylylsulfate reductase from2 desulfovibrio gigas
|
| 17 | c2zxiC_ |
|
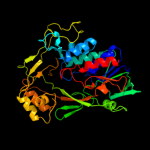 |
99.9 |
13 |
PDB header:fad-binding protein
Chain: C: PDB Molecule:trna uridine 5-carboxymethylaminomethyl
PDBTitle: structure of aquifex aeolicus gida in the form ii crystal
|
| 18 | c3g05B_ |
|
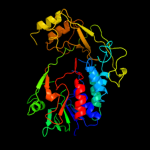 |
99.9 |
17 |
PDB header:rna binding protein
Chain: B: PDB Molecule:trna uridine 5-carboxymethylaminomethyl modification enzyme
PDBTitle: crystal structure of n-terminal domain (2-550) of e.coli mnmg
|
| 19 | c3cp8C_ |
|
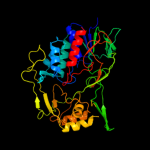 |
99.9 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:trna uridine 5-carboxymethylaminomethyl
PDBTitle: crystal structure of gida from chlorobium tepidum
|
| 20 | c3cesB_ |
|
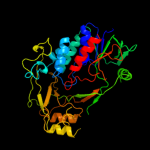 |
99.9 |
16 |
PDB header:rna binding protein
Chain: B: PDB Molecule:trna uridine 5-carboxymethylaminomethyl modification enzyme
PDBTitle: crystal structure of e.coli mnmg (gida), a highly-conserved trna2 modifying enzyme
|
| 21 | c3nlcA_ |
|
not modelled |
99.9 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vp0956;
PDBTitle: crystal structure of the vp0956 protein from vibrio parahaemolyticus.2 northeast structural genomics consortium target vpr147
|
| 22 | d1chua2 |
|
not modelled |
99.9 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 23 | d1qo8a2 |
|
not modelled |
99.9 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 24 | d1y0pa2 |
|
not modelled |
99.9 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 25 | d1d4ca2 |
|
not modelled |
99.8 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 26 | d2i0za1 |
|
not modelled |
99.8 |
31 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
| 27 | d1neka2 |
|
not modelled |
99.8 |
29 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 28 | c2eq8E_ |
|
not modelled |
99.8 |
29 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
|
| 29 | d3grsa1 |
|
not modelled |
99.8 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 30 | c1ojtA_ |
|
not modelled |
99.8 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:surface protein;
PDBTitle: structure of dihydrolipoamide dehydrogenase
|
| 31 | d1jnra2 |
|
not modelled |
99.8 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 32 | c1tytA_ |
|
not modelled |
99.8 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trypanothione reductase, oxidized form;
PDBTitle: crystal and molecular structure of crithidia fasciculata2 trypanothione reductase at 2.6 angstroms resolution
|
| 33 | c3o0hA_ |
|
not modelled |
99.8 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione reductase;
PDBTitle: crystal structure of glutathione reductase from bartonella henselae
|
| 34 | c1v59B_ |
|
not modelled |
99.7 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: crystal structure of yeast lipoamide dehydrogenase2 complexed with nad+
|
| 35 | c2c3dB_ |
|
not modelled |
99.7 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxopropyl-com reductase;
PDBTitle: 2.15 angstrom crystal structure of 2-ketopropyl coenzyme m2 oxidoreductase carboxylase with a coenzyme m disulfide3 bound at the active site
|
| 36 | c2w0hA_ |
|
not modelled |
99.7 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trypanothione reductase;
PDBTitle: x ray structure of leishmania infantum trypanothione2 reductase in complex with antimony and nadph
|
| 37 | c1hyuA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: crystal structure of intact ahpf
|
| 38 | c1lpfB_ |
|
not modelled |
99.7 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: three-dimensional structure of lipoamide dehydrogenase from2 pseudomonas fluorescens at 2.8 angstroms resolution.3 analysis of redox and thermostability properties
|
| 39 | c1geuA_ |
|
not modelled |
99.7 |
26 |
PDB header:oxidoreductase(flavoenzyme)
Chain: A: PDB Molecule:glutathione reductase;
PDBTitle: anatomy of an engineered nad-binding site
|
| 40 | c2eq7B_ |
|
not modelled |
99.7 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxoglutarate dehydrogenase e3 component;
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdo
|
| 41 | c1bwcA_ |
|
not modelled |
99.7 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (glutathione reductase);
PDBTitle: structure of human glutathione reductase complexed with ajoene2 inhibitor and subversive substrate
|
| 42 | c3r9uA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: thioredoxin-disulfide reductase from campylobacter jejuni.
|
| 43 | c3k7tB_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-hydroxy-l-nicotine oxidase;
PDBTitle: crystal structure of apo-form 6-hydroxy-l-nicotine oxidase,2 crystal form p3121
|
| 44 | c1ndaD_ |
|
not modelled |
99.7 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:trypanothione oxidoreductase;
PDBTitle: the structure of trypanosoma cruzi trypanothione reductase2 in the oxidized and nadph reduced state
|
| 45 | c3dgzA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase 2;
PDBTitle: crystal structure of mouse mitochondrial thioredoxin reductase, c-2 terminal 3-residue truncation
|
| 46 | c3rhaA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putrescine oxidase;
PDBTitle: the crystal structure of oxidoreductase from arthrobacter aurescens
|
| 47 | c1s3bB_ |
|
not modelled |
99.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine oxidase [flavin-containing] b;
PDBTitle: crystal structure of maob in complex with n-methyl-n-2 propargyl-1(r)-aminoindan
|
| 48 | c2v6oA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin glutathione reductase;
PDBTitle: structure of schistosoma mansoni thioredoxin-glutathione2 reductase (smtgr)
|
| 49 | c3fbsB_ |
|
not modelled |
99.7 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of the oxidoreductase from agrobacterium2 tumefaciens
|
| 50 | c1zkqA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase 2, mitochondrial;
PDBTitle: crystal structure of mouse thioredoxin reductase type 2
|
| 51 | c2zbwA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of thioredoxin reductase-like protein from thermus2 thermophilus hb8
|
| 52 | c1zx9A_ |
|
not modelled |
99.7 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mercuric reductase;
PDBTitle: crystal structure of tn501 mera
|
| 53 | c3urhB_ |
|
not modelled |
99.7 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of a dihydrolipoamide dehydrogenase from2 sinorhizobium meliloti 1021
|
| 54 | d1kf6a2 |
|
not modelled |
99.7 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 55 | c2a8xA_ |
|
not modelled |
99.7 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of lipoamide dehydrogenase from2 mycobacterium tuberculosis
|
| 56 | d3lada1 |
|
not modelled |
99.7 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 57 | c2qaeA_ |
|
not modelled |
99.7 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure analysis of trypanosoma cruzi lipoamide2 dehydrogenase
|
| 58 | c1yvvB_ |
|
not modelled |
99.7 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine oxidase, flavin-containing;
PDBTitle: x-ray structurure of p. syringae q888a4 oxidoreductase at2 resolution 2.5a. northeast structural genomics consortium3 target psr10.
|
| 59 | d1lpfa1 |
|
not modelled |
99.7 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 60 | c2yg4B_ |
|
not modelled |
99.7 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putrescine oxidase;
PDBTitle: structure-based redesign of cofactor binding in putrescine2 oxidase: wild type bound to putrescine
|
| 61 | c2nvkX_ |
|
not modelled |
99.7 |
29 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of thioredoxin reductase from drosophila2 melanogaster
|
| 62 | c3f8rD_ |
|
not modelled |
99.7 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:thioredoxin reductase (trxb-3);
PDBTitle: crystal structure of sulfolobus solfataricus thioredoxin2 reductase b3 in complex with two nadp molecules
|
| 63 | c1onfA_ |
|
not modelled |
99.7 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione reductase;
PDBTitle: crystal structure of plasmodium falciparum glutathione reductase
|
| 64 | d1v59a1 |
|
not modelled |
99.7 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 65 | c3l8kB_ |
|
not modelled |
99.7 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of a dihydrolipoyl dehydrogenase from2 sulfolobus solfataricus
|
| 66 | c2r9zB_ |
|
not modelled |
99.7 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione amide reductase;
PDBTitle: glutathione amide reductase from chromatium gracile
|
| 67 | c3ctyA_ |
|
not modelled |
99.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of t. acidophilum thioredoxin reductase
|
| 68 | d1h6va1 |
|
not modelled |
99.7 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 69 | c2gmhA_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:electron transfer flavoprotein-ubiquinone
PDBTitle: structure of porcine electron transfer flavoprotein-2 ubiquinone oxidoreductase in complexed with ubiquinone
|
| 70 | c1dxlC_ |
|
not modelled |
99.7 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: dihydrolipoamide dehydrogenase of glycine decarboxylase2 from pisum sativum
|
| 71 | d1dxla1 |
|
not modelled |
99.6 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 72 | c2hqmB_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione reductase;
PDBTitle: crystal structure of glutathione reductase glr1 from the yeast2 saccharomyces cerevisiae
|
| 73 | d1ojta1 |
|
not modelled |
99.6 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 74 | c3lzxB_ |
|
not modelled |
99.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin--nadp reductase 2;
PDBTitle: crystal structure of ferredoxin-nadp+ oxidoreductase from bacillus2 subtilis (form ii)
|
| 75 | c2q0lA_ |
|
not modelled |
99.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: helicobacter pylori thioredoxin reductase reduced by sodium dithionite2 in complex with nadp+
|
| 76 | c1f6mF_ |
|
not modelled |
99.6 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of a complex between thioredoxin2 reductase, thioredoxin, and the nadp+ analog, aadp+
|
| 77 | c2xagA_ |
|
not modelled |
99.6 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: crystal structure of lsd1-corest in complex with para-bromo-2 (-)-trans-2-phenylcyclopropyl-1-amine
|
| 78 | c2v1dA_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase/repressor
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: structural basis of lsd1-corest selectivity in histone h32 recognition
|
| 79 | d1aoga1 |
|
not modelled |
99.6 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 80 | c3g5rA_ |
|
not modelled |
99.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:methylenetetrahydrofolate--trna-(uracil-5-)-
PDBTitle: crystal structure of thermus thermophilus trmfo in complex with2 tetrahydrofolate
|
| 81 | c2ivdA_ |
|
not modelled |
99.6 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: structure of protoporphyrinogen oxidase from myxococcus2 xanthus with acifluorfen
|
| 82 | c1zmcG_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of human dihydrolipoamide dehydrogenase2 complexed to nad+
|
| 83 | c2hkoA_ |
|
not modelled |
99.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: crystal structure of lsd1
|
| 84 | c1ebdB_ |
|
not modelled |
99.6 |
15 |
PDB header:complex (oxidoreductase/transferase)
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: dihydrolipoamide dehydrogenase complexed with the binding2 domain of the dihydrolipoamide acetylase
|
| 85 | c3d8xB_ |
|
not modelled |
99.6 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase 1;
PDBTitle: crystal structure of saccharomyces cerevisiae nadph dependent2 thioredoxin reductase 1
|
| 86 | c2cfyB_ |
|
not modelled |
99.6 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase 1;
PDBTitle: crystal structure of human thioredoxin reductase 1
|
| 87 | c3ic9D_ |
|
not modelled |
99.6 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: the structure of dihydrolipoamide dehydrogenase from colwellia2 psychrerythraea 34h.
|
| 88 | c3i6dA_ |
|
not modelled |
99.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: crystal structure of ppo from bacillus subtilis with af
|
| 89 | d1ebda1 |
|
not modelled |
99.6 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 90 | d2gmha1 |
|
not modelled |
99.6 |
18 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 91 | c3ab1B_ |
|
not modelled |
99.6 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin--nadp reductase;
PDBTitle: crystal structure of ferredoxin nadp+ oxidoreductase
|
| 92 | c1h83A_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyamine oxidase;
PDBTitle: structure of polyamine oxidase in complex with2 1,8-diaminooctane
|
| 93 | c1vdcA_ |
|
not modelled |
99.6 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dependent thioredoxin reductase;
PDBTitle: structure of nadph dependent thioredoxin reductase
|
| 94 | d1lvla1 |
|
not modelled |
99.6 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 95 | c2a87A_ |
|
not modelled |
99.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of m. tuberculosis thioredoxin reductase
|
| 96 | c3atrA_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:conserved archaeal protein;
PDBTitle: geranylgeranyl reductase (ggr) from sulfolobus acidocaldarius co-2 crystallized with its ligand
|
| 97 | d1ryia1 |
|
not modelled |
99.6 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 98 | c1fl2A_ |
|
not modelled |
99.6 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: catalytic core component of the alkylhydroperoxide reductase ahpf from2 e.coli
|
| 99 | c3nksA_ |
|
not modelled |
99.6 |
14 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: structure of human protoporphyrinogen ix oxidase
|
| 100 | c1sezA_ |
|
not modelled |
99.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase, mitochondrial;
PDBTitle: crystal structure of protoporphyrinogen ix oxidase
|
| 101 | c1lvlA_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: the refined structure of pseudomonas putida lipoamide dehydrogenase2 complexed with nad+ at 2.45 angstroms resolution
|
| 102 | c3kd9B_ |
|
not modelled |
99.6 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:coenzyme a disulfide reductase;
PDBTitle: crystal structure of pyridine nucleotide disulfide oxidoreductase from2 pyrococcus horikoshii
|
| 103 | c3cgdB_ |
|
not modelled |
99.5 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyridine nucleotide-disulfide oxidoreductase, class i;
PDBTitle: pyridine nucleotide complexes with bacillus anthracis coenzyme a-2 disulfide reductase: a structural analysis of dual nad(p)h3 specificity
|
| 104 | c3kljA_ |
|
not modelled |
99.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad(fad)-dependent dehydrogenase, nirb-family (n-terminal
PDBTitle: crystal structure of nadh:rubredoxin oxidoreductase from clostridium2 acetobutylicum
|
| 105 | d2bs2a2 |
|
not modelled |
99.5 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 106 | d1feca1 |
|
not modelled |
99.5 |
27 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 107 | c2r4jA_ |
|
not modelled |
99.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aerobic glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of escherichia coli semet substituted2 glycerol-3-phosphate dehydrogenase in complex with dhap
|
| 108 | c3qj4A_ |
|
not modelled |
99.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:renalase;
PDBTitle: crystal structure of human renalase (isoform 1)
|
| 109 | c1q1wA_ |
|
not modelled |
99.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putidaredoxin reductase;
PDBTitle: crystal structure of putidaredoxin reductase from2 pseudomonas putida
|
| 110 | c2cduB_ |
|
not modelled |
99.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph oxidase;
PDBTitle: the crystal structure of water-forming nad(p)h oxidase from2 lactobacillus sanfranciscensis
|
| 111 | d2gf3a1 |
|
not modelled |
99.5 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 112 | c2gr2A_ |
|
not modelled |
99.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
| 113 | c1gthD_ |
|
not modelled |
99.5 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydropyrimidine dehydrogenase;
PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
|
| 114 | c3jskN_ |
|
not modelled |
99.5 |
20 |
PDB header:biosynthetic protein
Chain: N: PDB Molecule:cypbp37 protein;
PDBTitle: thiazole synthase from neurospora crassa
|
| 115 | c1xdiA_ |
|
not modelled |
99.5 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:rv3303c-lpda;
PDBTitle: crystal structure of lpda (rv3303c) from mycobacterium tuberculosis
|
| 116 | c3lxdA_ |
|
not modelled |
99.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: crystal structure of ferredoxin reductase arr from novosphingobium2 aromaticivorans
|
| 117 | c1gv4A_ |
|
not modelled |
99.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:programed cell death protein 8;
PDBTitle: murine apoptosis-inducing factor (aif)
|
| 118 | c3da1A_ |
|
not modelled |
99.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: x-ray structure of the glycerol-3-phosphate dehydrogenase2 from bacillus halodurans complexed with fad. northeast3 structural genomics consortium target bhr167.
|
| 119 | d1rp0a1 |
|
not modelled |
99.4 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Thi4-like |
| 120 | c2vvlD_ |
|
not modelled |
99.4 |
27 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:monoamine oxidase n;
PDBTitle: the structure of mao-n-d3, a variant of monoamine oxidase2 from aspergillus niger.
|

