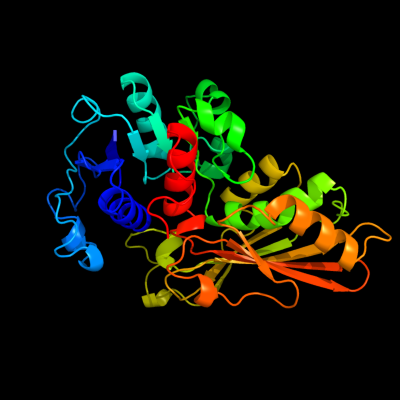
| 1 | c2g17A_
|
|
 |
100.0 |
90 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: the structure of n-acetyl-gamma-glutamyl-phosphate reductase from2 salmonella typhimurium.
|
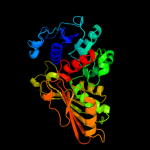
| 2 | c2q49B_
|
|
 |
100.0 |
37 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at2g19940
|
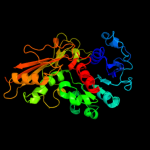
| 3 | c2ozpA_
|
|
 |
100.0 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (ttha1904) from thermus thermophilus
|
| 4 | c2i3aD_
|
|
 |
100.0 |
38 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (rv1652) from mycobacterium tuberculosis
|
| 5 | c1vknC_
|
|
 |
100.0 |
37 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (tm1782) from thermotoga maritima at 1.80 a resolution
|
| 6 | c2ep5B_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:350aa long hypothetical aspartate-semialdehyde
PDBTitle: structural study of project id st1242 from sulfolobus tokodaii strain7
|
| 7 | c1mb4B_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 from vibrio cholerae with nadp and s-methyl-l-cysteine3 sulfoxide
|
| 8 | c3hskB_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 with nadp from candida albicans
|
| 9 | c1ys4A_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: structure of aspartate-semialdehyde dehydrogenase from2 methanococcus jannaschii
|
| 10 | c1t4bB_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: 1.6 angstrom structure of esherichia coli aspartate-2 semialdehyde dehydrogenase.
|
| 11 | c2gz3D_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aspartate beta-semialdehyde dehydrogenase;
PDBTitle: structure of aspartate semialdehyde dehydrogenase (asadh) from2 streptococcus pneumoniae complexed with nadp and aspartate-3 semialdehyde
|
| 12 | c3uw3A_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of an aspartate-semialdehyde dehydrogenase from2 burkholderia thailandensis
|
| 13 | c3kubA_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semi-aldehyde dehydrogenase complexed2 with glycerol and phosphate of mycobacterium tuberculosis h37rv
|
| 14 | c2yv3B_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase from thermus2 thermophilus hb8
|
| 15 | c2qz9B_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 ii from vibrio cholerae
|
| 16 | c2hjsA_
|
|
 |
100.0 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:usg-1 protein homolog;
PDBTitle: the structure of a probable aspartate-semialdehyde dehydrogenase from2 pseudomonas aeruginosa
|
| 17 | c2gd1P_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase(aldehyde(d)-nad(a))
Chain: P: PDB Molecule:apo-d-glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: coenzyme-induced conformational changes in glyceraldehyde-3-2 phosphate dehydrogenase from bacillus stearothermophillus
|
| 18 | c3sthA_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 toxoplasma gondii
|
| 19 | c1s7cA_
|
|
 |
100.0 |
14 |
PDB header:structural genomics, oxidoreductase
Chain: A: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase a;
PDBTitle: crystal structure of mes buffer bound form of glyceraldehyde 3-2 phosphate dehydrogenase from escherichia coli
|
| 20 | d2g17a1
|
|
 |
100.0 |
78 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 21 | c3cieC_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
|
| 22 | d2q49a1 |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 23 | c1cerC_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase (aldehyde(d)-nad(a))
Chain: C: PDB Molecule:holo-d-glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: determinants of enzyme thermostability observed in the2 molecular structure of thermus aquaticus d-glyceraldehyde-3 3-phosphate dehydrogenase at 2.5 angstroms resolution
|
| 24 | c1cf2Q_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:protein (glyceraldehyde-3-phosphate
PDBTitle: three-dimensional structure of d-glyceraldehyde-3-phosphate2 dehydrogenase from the hyperthermophilic archaeon3 methanothermus fervidus
|
| 25 | c1hdgO_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase (aldehy(d)-nad(a))
Chain: O: PDB Molecule:holo-d-glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
|
| 26 | c1rm4O_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase a;
PDBTitle: crystal structure of recombinant photosynthetic glyceraldehyde-3-2 phosphate dehydrogenase a4 isoform, complexed with nadp
|
| 27 | d2cvoa1 |
|
not modelled |
100.0 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 28 | c1b7gO_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:protein (glyceraldehyde 3-phosphate dehydrogenase);
PDBTitle: glyceraldehyde 3-phosphate dehydrogenase
|
| 29 | c2d2iO_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-2 phosphate dehydrogenase from synechococcus sp. complexed3 with nadp+
|
| 30 | c2pkrI_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase aor;
PDBTitle: crystal structure of (a+cte)4 chimeric form of2 photosyntetic glyceraldehyde-3-phosphate dehydrogenase,3 complexed with nadp
|
| 31 | c3h9eA_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:
PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
|
| 32 | c2yyyB_ |
|
not modelled |
100.0 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase
|
| 33 | d1vkna1 |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 34 | c2x5kO_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:d-erythrose-4-phosphate dehydrogenase;
PDBTitle: structure of an active site mutant of the d-erythrose-4-phosphate2 dehydrogenase from e. coli
|
| 35 | c2i5pO_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase 1;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase isoform 1 from k. marxianus
|
| 36 | c3docD_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: crystal structure of trka glyceraldehyde-3-phosphate2 dehydrogenase from brucella melitensis
|
| 37 | c2b4rQ_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
|
| 38 | c2ep7B_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
|
| 39 | c3hq4R_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: R: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase 1;
PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
|
| 40 | c2czcD_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 pyrococcus horikoshii ot3
|
| 41 | c3hjaB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase from borrelia burgdorferi
|
| 42 | c1ihxD_ |
|
not modelled |
100.0 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: crystal structure of two d-glyceraldehyde-3-phosphate2 dehydrogenase complexes: a case of asymmetry
|
| 43 | c1obfO_ |
|
not modelled |
100.0 |
14 |
PDB header:glycolytic pathway
Chain: O: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.73 resolution.
|
| 44 | d2g17a2 |
|
not modelled |
100.0 |
92 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 45 | d2cvoa2 |
|
not modelled |
100.0 |
38 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 46 | c1i32D_ |
|
not modelled |
100.0 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: leishmania mexicana glyceraldehyde-3-phosphate2 dehydrogenase in complex with inhibitors
|
| 47 | d1vkna2 |
|
not modelled |
100.0 |
39 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 48 | d2q49a2 |
|
not modelled |
100.0 |
40 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 49 | d2gz1a1 |
|
not modelled |
100.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 50 | d2hjsa2 |
|
not modelled |
100.0 |
20 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 51 | d2hjsa1 |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 52 | d1pqua2 |
|
not modelled |
100.0 |
24 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 53 | d1t4ba2 |
|
not modelled |
100.0 |
26 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 54 | d2gz1a2 |
|
not modelled |
100.0 |
24 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 55 | d1mb4a2 |
|
not modelled |
100.0 |
24 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 56 | d1t4ba1 |
|
not modelled |
100.0 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 57 | d1pqua1 |
|
not modelled |
100.0 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 58 | d1mb4a1 |
|
not modelled |
100.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 59 | d1hdgo2 |
|
not modelled |
99.9 |
14 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 60 | d1cf2o1 |
|
not modelled |
99.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 61 | d1rm4a2 |
|
not modelled |
99.9 |
8 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 62 | d1dssg1 |
|
not modelled |
99.9 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 63 | d1u8fo2 |
|
not modelled |
99.9 |
12 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 64 | d3cmco2 |
|
not modelled |
99.9 |
12 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 65 | d2b4ro2 |
|
not modelled |
99.9 |
13 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 66 | d1gado2 |
|
not modelled |
99.9 |
14 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 67 | d1i32a2 |
|
not modelled |
99.9 |
11 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 68 | d1ggaa2 |
|
not modelled |
99.8 |
8 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 69 | d3gpdg2 |
|
not modelled |
99.8 |
13 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 70 | d2pkqo2 |
|
not modelled |
99.8 |
14 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 71 | d2g82a2 |
|
not modelled |
99.8 |
15 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 72 | d1dssg2 |
|
not modelled |
99.8 |
11 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 73 | d1gado1 |
|
not modelled |
99.8 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 74 | d1k3ta2 |
|
not modelled |
99.8 |
9 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 75 | d3cmco1 |
|
not modelled |
99.8 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 76 | d1obfo2 |
|
not modelled |
99.8 |
11 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 77 | d1j0xo1 |
|
not modelled |
99.8 |
8 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 78 | d1rm4a1 |
|
not modelled |
99.8 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 79 | d1u8fo1 |
|
not modelled |
99.8 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 80 | d2g82a1 |
|
not modelled |
99.8 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 81 | d1ggaa1 |
|
not modelled |
99.8 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 82 | d1vc2a1 |
|
not modelled |
99.7 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 83 | d1k3ta1 |
|
not modelled |
99.7 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 84 | d1hdgo1 |
|
not modelled |
99.7 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 85 | d1i32a1 |
|
not modelled |
99.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 86 | d2pkqo1 |
|
not modelled |
99.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 87 | d3gpdg1 |
|
not modelled |
99.7 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 88 | d2b4ro1 |
|
not modelled |
99.6 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 89 | d1obfo1 |
|
not modelled |
99.6 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 90 | c1drwA_ |
|
not modelled |
99.2 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
| 91 | d2czca2 |
|
not modelled |
99.2 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 92 | c1nvmB_ |
|
not modelled |
99.1 |
17 |
PDB header:lyase/oxidoreductase
Chain: B: PDB Molecule:acetaldehyde dehydrogenase (acylating);
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 93 | c3ijpA_ |
|
not modelled |
99.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase from2 bartonella henselae at 2.0a resolution
|
| 94 | c3ic5A_ |
|
not modelled |
99.1 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 95 | c3dapB_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:diaminopimelic acid dehydrogenase;
PDBTitle: c. glutamicum dap dehydrogenase in complex with nadp+ and2 the inhibitor 5s-isoxazoline
|
| 96 | d1diha1 |
|
not modelled |
99.0 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 97 | d1f06a1 |
|
not modelled |
98.9 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 98 | c2z2vA_ |
|
not modelled |
98.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1688;
PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
|
| 99 | d1nvmb1 |
|
not modelled |
98.9 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 100 | d1b7go1 |
|
not modelled |
98.9 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 101 | c2ho3D_ |
|
not modelled |
98.8 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
| 102 | d1cf2o2 |
|
not modelled |
98.8 |
13 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 103 | c3bioB_ |
|
not modelled |
98.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase (gfo/idh/moca family member) from2 porphyromonas gingivalis w83
|
| 104 | c3fhlC_ |
|
not modelled |
98.7 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from bacteroides2 fragilis nctc 9343
|
| 105 | c1e5lA_ |
|
not modelled |
98.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
| 106 | d1b7go2 |
|
not modelled |
98.6 |
16 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 107 | c2dt5A_ |
|
not modelled |
98.6 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:at-rich dna-binding protein;
PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
|
| 108 | c2axqA_ |
|
not modelled |
98.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 109 | d1q0qa2 |
|
not modelled |
98.6 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 110 | d2dt5a2 |
|
not modelled |
98.6 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Transcriptional repressor Rex, C-terminal domain |
| 111 | c3m2tA_ |
|
not modelled |
98.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
|
| 112 | c2q4eB_ |
|
not modelled |
98.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
| 113 | c2ixaA_ |
|
not modelled |
98.5 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 114 | c3f4lF_ |
|
not modelled |
98.5 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative oxidoreductase yhhx;
PDBTitle: crystal structure of a probable oxidoreductase yhhx in2 triclinic form. northeast structural genomics target er647
|
| 115 | c3db2C_ |
|
not modelled |
98.5 |
8 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 116 | c3e18A_ |
|
not modelled |
98.5 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 117 | c3do5A_ |
|
not modelled |
98.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoserine dehydrogenase;
PDBTitle: crystal structure of putative homoserine dehydrogenase (np_069768.1)2 from archaeoglobus fulgidus at 2.20 a resolution
|
| 118 | d2czca1 |
|
not modelled |
98.5 |
14 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 119 | c3a14B_ |
|
not modelled |
98.5 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
|
| 120 | c3euwB_ |
|
not modelled |
98.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|