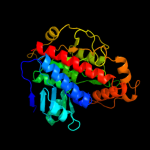
| 1 | d1vkma_ |
|
 |
100.0 |
38 |
Fold:Indigoidine synthase A-like
Superfamily:Indigoidine synthase A-like
Family:Indigoidine synthase A-like |
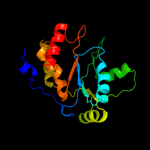
| 2 | c1otpA_ |
|
 |
93.4 |
21 |
PDB header:phosphorylase
Chain: A: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
|
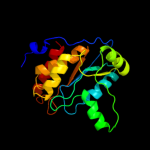
| 3 | c2j0fC_ |
|
 |
93.1 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural basis for non-competitive product inhibition in2 human thymidine phosphorylase: implication for drug design
|
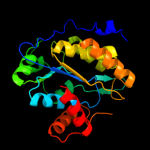
| 4 | c1brwB_ |
|
 |
93.1 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyrimidine nucleoside phosphorylase);
PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
|
| 5 | c2dsjA_ |
|
 |
92.3 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside (thymidine) phosphorylase;
PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
|
| 6 | c3h5qA_ |
|
 |
91.6 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside phosphorylase;
PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
|
| 7 | c1zfjA_ |
|
 |
91.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 8 | d1s2wa_ |
|
 |
90.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 9 | d1zfja1 |
|
 |
86.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 10 | d1muma_ |
|
 |
83.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 11 | c1khdD_ |
|
 |
83.0 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure analysis of the anthranilate2 phosphoribosyltransferase from erwinia carotovora at 1.93 resolution (current name, pectobacterium carotovorum)
|
| 12 | c2fr1A_ |
|
 |
81.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:erythromycin synthase, eryai;
PDBTitle: the first ketoreductase of the erythromycin synthase2 (crystal form 2)
|
| 13 | d2elca2 |
|
 |
80.4 |
27 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 14 | c1v8gB_ |
|
 |
79.7 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure of anthranilate phosphoribosyltransferase2 (trpd) from thermus thermophilus hb8
|
| 15 | c1o17A_ |
|
 |
78.1 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
|
| 16 | c2vz8B_ |
|
 |
77.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase;
PDBTitle: crystal structure of mammalian fatty acid synthase
|
| 17 | c3mjsA_ |
|
 |
76.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:amphb;
PDBTitle: structure of a-type ketoreductases from modular polyketide synthase
|
| 18 | d1jvna1 |
|
 |
74.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 19 | c2z5lA_ |
|
 |
73.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:tylactone synthase starter module and modules 1
PDBTitle: the first ketoreductase of the tylosin pks
|
| 20 | d1r7ha_ |
|
 |
68.3 |
20 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 21 | c2hjpA_ |
|
not modelled |
67.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 22 | c1vquB_ |
|
not modelled |
65.8 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase 2;
PDBTitle: crystal structure of anthranilate phosphoribosyltransferase 22 (17130499) from nostoc sp. at 1.85 a resolution
|
| 23 | c3fmfA_ |
|
not modelled |
65.8 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of mycobacterium tuberculosis dethiobiotin2 synthetase complexed with 7,8 diaminopelargonic acid carbamate
|
| 24 | c3n2oA_ |
|
not modelled |
64.6 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: x-ray crystal structure of arginine decarboxylase complexed with2 arginine from vibrio vulnificus
|
| 25 | c3khsB_ |
|
not modelled |
64.2 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
|
| 26 | c3lyhB_ |
|
not modelled |
63.3 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:cobalamin (vitamin b12) biosynthesis cbix protein;
PDBTitle: crystal structure of putative cobalamin (vitamin b12) biosynthesis2 cbix protein (yp_958415.1) from marinobacter aquaeolei vt8 at 1.60 a3 resolution
|
| 27 | d1mzha_ |
|
not modelled |
63.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 28 | c3qp9C_ |
|
not modelled |
59.7 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:type i polyketide synthase pikaii;
PDBTitle: the structure of a c2-type ketoreductase from a modular polyketide2 synthase
|
| 29 | d1uoua2 |
|
not modelled |
59.4 |
23 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 30 | c3nzpA_ |
|
not modelled |
59.3 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:arginine decarboxylase;
PDBTitle: crystal structure of the biosynthetic arginine decarboxylase spea from2 campylobacter jejuni, northeast structural genomics consortium target3 br53
|
| 31 | c1zlpA_ |
|
not modelled |
59.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 32 | d1m3ua_ |
|
not modelled |
58.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 33 | c3ih1A_ |
|
not modelled |
57.6 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 34 | c3ffsC_ |
|
not modelled |
54.6 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: the crystal structure of cryptosporidium parvum inosine-5'-2 monophosphate dehydrogenase
|
| 35 | d1brwa2 |
|
not modelled |
54.3 |
27 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 36 | c1ypfB_ |
|
not modelled |
51.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gmp reductase;
PDBTitle: crystal structure of guac (ba5705) from bacillus anthracis at 1.8 a2 resolution
|
| 37 | c3ggsA_ |
|
not modelled |
49.3 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: human purine nucleoside phosphorylase double mutant e201q,n243d2 complexed with 2-fluoro-2'-deoxyadenosine
|
| 38 | d1t9ba1 |
|
not modelled |
47.9 |
28 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 39 | d1f0ka_ |
|
not modelled |
47.7 |
16 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
| 40 | c1yadD_ |
|
not modelled |
47.6 |
24 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein teni;
PDBTitle: structure of teni from bacillus subtilis
|
| 41 | d2ihta1 |
|
not modelled |
47.1 |
24 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 42 | c3cf4G_ |
|
not modelled |
45.1 |
15 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:acetyl-coa decarboxylase/synthase epsilon subunit;
PDBTitle: structure of the codh component of the m. barkeri acds complex
|
| 43 | c2bibA_ |
|
not modelled |
45.1 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:teichoic acid phosphorylcholine esterase/ choline binding
PDBTitle: crystal structure of the complete modular teichioic acid2 phosphorylcholine esterase pce (cbpe) from streptococcus3 pneumoniae
|
| 44 | d1xm3a_ |
|
not modelled |
45.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 45 | d1khda2 |
|
not modelled |
44.9 |
21 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 46 | c1jvnB_ |
|
not modelled |
43.8 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 47 | c3mioA_ |
|
not modelled |
42.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:3,4-dihydroxy-2-butanone 4-phosphate synthase;
PDBTitle: crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase2 domain from mycobacterium tuberculosis at ph 6.00
|
| 48 | d3bgsa1 |
|
not modelled |
41.1 |
24 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 49 | d1ujqa_ |
|
not modelled |
40.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 50 | d3pnpa_ |
|
not modelled |
39.7 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 51 | d1zpda1 |
|
not modelled |
39.2 |
5 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 52 | c3guzB_ |
|
not modelled |
37.8 |
26 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: structural and substrate-binding studies of pantothenate2 synthenate (ps)provide insights into homotropic inhibition3 by pantoate in ps's
|
| 53 | c2qr6A_ |
|
not modelled |
37.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:imp dehydrogenase/gmp reductase;
PDBTitle: crystal structure of imp dehydrogenase/gmp reductase-like protein2 (np_599840.1) from corynebacterium glutamicum atcc 13032 kitasato at3 1.50 a resolution
|
| 54 | c2agkA_ |
|
not modelled |
37.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: structure of s. cerevisiae his6 protein
|
| 55 | d1h5ya_ |
|
not modelled |
36.1 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 56 | c1jd7A_ |
|
not modelled |
35.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure analysis of the mutant k300r of2 pseudoalteromonas haloplanctis alpha-amylase
|
| 57 | d1wdia_ |
|
not modelled |
34.9 |
27 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
| 58 | c2qmoA_ |
|
not modelled |
34.5 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of dethiobiotin synthetase (biod) from helicobacter2 pylori
|
| 59 | d1n57a_ |
|
not modelled |
34.4 |
33 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:DJ-1/PfpI |
| 60 | d2ae2a_ |
|
not modelled |
34.2 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 61 | d1v77a_ |
|
not modelled |
33.9 |
10 |
Fold:7-stranded beta/alpha barrel
Superfamily:PHP domain-like
Family:RNase P subunit p30 |
| 62 | c2yvqA_ |
|
not modelled |
33.5 |
58 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 63 | c3eooL_ |
|
not modelled |
33.4 |
17 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 64 | c2xvzA_ |
|
not modelled |
32.8 |
11 |
PDB header:metal binding protein
Chain: A: PDB Molecule:chelatase, putative;
PDBTitle: cobalt chelatase cbik (periplasmatic) from desulvobrio2 vulgaris hildenborough (co-crystallized with cobalt)
|
| 65 | c3rggD_ |
|
not modelled |
32.6 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase, pure protein;
PDBTitle: crystal structure of treponema denticola pure bound to air
|
| 66 | c3chvA_ |
|
not modelled |
32.0 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
| 67 | c3n6rK_ |
|
not modelled |
31.5 |
22 |
PDB header:ligase
Chain: K: PDB Molecule:propionyl-coa carboxylase, alpha subunit;
PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
| 68 | c3nxkE_ |
|
not modelled |
31.2 |
22 |
PDB header:hydrolase
Chain: E: PDB Molecule:cytoplasmic l-asparaginase;
PDBTitle: crystal structure of probable cytoplasmic l-asparaginase from2 campylobacter jejuni
|
| 69 | c3e02A_ |
|
not modelled |
29.8 |
23 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
| 70 | d1u11a_ |
|
not modelled |
29.6 |
10 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 71 | c3b40A_ |
|
not modelled |
29.5 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable dipeptidase;
PDBTitle: crystal structure of the probable dipeptidase pvdm from2 pseudomonas aeruginosa
|
| 72 | d1ebda3 |
|
not modelled |
28.5 |
16 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family:FAD/NAD-linked reductases, dimerisation (C-terminal) domain |
| 73 | c2a7rD_ |
|
not modelled |
27.2 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gmp reductase 2;
PDBTitle: crystal structure of human guanosine monophosphate2 reductase 2 (gmpr2)
|
| 74 | c3nzqB_ |
|
not modelled |
27.2 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
|
| 75 | c2fw9A_ |
|
not modelled |
26.5 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
|
| 76 | c2vedA_ |
|
not modelled |
25.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:membrane protein capa1, protein tyrosine kinase;
PDBTitle: crystal structure of the chimerical mutant capabk55m2 protein
|
| 77 | d2tpta2 |
|
not modelled |
25.2 |
21 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 78 | d1g2oa_ |
|
not modelled |
24.6 |
24 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 79 | d2ji7a1 |
|
not modelled |
24.5 |
23 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 80 | c3e49A_ |
|
not modelled |
24.0 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849 with a tim barrel fold;
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
|
| 81 | d1ovma1 |
|
not modelled |
23.2 |
6 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 82 | d1q6za1 |
|
not modelled |
22.9 |
14 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 83 | c3majA_ |
|
not modelled |
22.8 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna processing chain a;
PDBTitle: crystal structure of putative dna processing protein dpra from2 rhodopseudomonas palustris cga009
|
| 84 | d1wmaa1 |
|
not modelled |
22.6 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 85 | c1a31A_ |
|
not modelled |
22.6 |
14 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:protein (topoisomerase i);
PDBTitle: human reconstituted dna topoisomerase i in covalent complex2 with a 22 base pair dna duplex
|
| 86 | c3ceuA_ |
|
not modelled |
22.6 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamine phosphate pyrophosphorylase2 (bt_0647) from bacteroides thetaiotaomicron. northeast3 structural genomics consortium target btr268
|
| 87 | d2adra2 |
|
not modelled |
22.4 |
28 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 88 | d1v58a2 |
|
not modelled |
22.3 |
22 |
Fold:Cystatin-like
Superfamily:DsbC/DsbG N-terminal domain-like
Family:DsbC/DsbG N-terminal domain-like |
| 89 | d3grsa3 |
|
not modelled |
22.3 |
20 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family:FAD/NAD-linked reductases, dimerisation (C-terminal) domain |
| 90 | c3oixA_ |
|
not modelled |
22.2 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative dihydroorotate dehydrogenase; dihydroorotate
PDBTitle: crystal structure of the putative dihydroorotate dehydrogenase from2 streptococcus mutans
|
| 91 | d1vrda1 |
|
not modelled |
22.1 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 92 | c1mg7B_ |
|
not modelled |
21.9 |
21 |
PDB header:gene regulation
Chain: B: PDB Molecule:early switch protein xol-1 2.2k splice form;
PDBTitle: crystal structure of xol-1
|
| 93 | d1k4ia_ |
|
not modelled |
21.8 |
23 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 94 | d1pkla2 |
|
not modelled |
21.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 95 | c3o2sB_ |
|
not modelled |
21.6 |
33 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72 complex
|
| 96 | c3o2qB_ |
|
not modelled |
21.5 |
30 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
|
| 97 | d1ozha1 |
|
not modelled |
21.3 |
20 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 98 | d1wraa1 |
|
not modelled |
21.0 |
13 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Pce catalytic domain-like |
| 99 | d2dasa1 |
|
not modelled |
20.9 |
32 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:TRASH domain |
| 100 | d1j6ua1 |
|
not modelled |
20.8 |
12 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 101 | c2dasA_ |
|
not modelled |
20.6 |
32 |
PDB header:metal transport
Chain: A: PDB Molecule:zinc finger mym-type protein 5;
PDBTitle: solution structure of trash domain of zinc finger mym-type2 protein 5
|
| 102 | c3labA_ |
|
not modelled |
20.5 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate)
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
| 103 | c3lu2B_ |
|
not modelled |
20.4 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2462 protein;
PDBTitle: structure of lmo2462, a listeria monocytogenes amidohydrolase family2 putative dipeptidase
|
| 104 | d1o17a2 |
|
not modelled |
20.4 |
18 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 105 | d1aoga3 |
|
not modelled |
20.3 |
18 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family:FAD/NAD-linked reductases, dimerisation (C-terminal) domain |
| 106 | c3tovB_ |
|
not modelled |
20.1 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase family 9;
PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
|
| 107 | d1tksa_ |
|
not modelled |
20.1 |
21 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |

