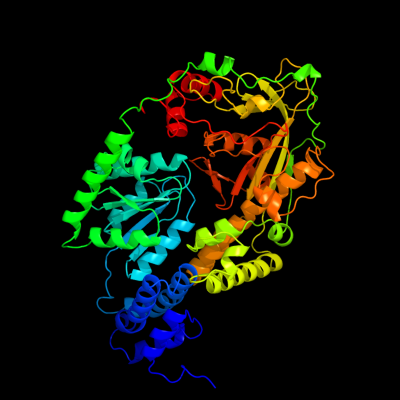
| 1 | c1k98A_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
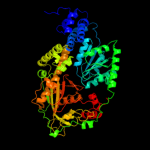
| 2 | c3bolB_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
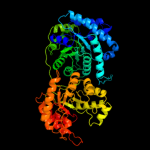
| 3 | d1mska_
|
|
 |
100.0 |
100 |
Fold:Methionine synthase activation domain-like
Superfamily:Methionine synthase activation domain-like
Family:Methionine synthase SAM-binding domain |
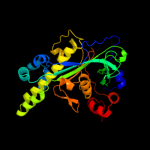
| 4 | c2o2kA_
|
|
 |
100.0 |
49 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: crystal structure of the activation domain of human2 methionine synthase isoform/mutant d963e/k1071n
|
| 5 | d1umya_
|
|
 |
100.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 6 | c3k13A_
|
|
 |
100.0 |
64 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 7 | d3bofa2
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 8 | d1lt7a_
|
|
 |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 9 | c2yciX_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 10 | d3bofa1
|
|
 |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 11 | c1bmtB_
|
|
 |
100.0 |
100 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
| 12 | d1f6ya_
|
|
 |
100.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 13 | c2i2xD_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
| 14 | c3ezxA_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
| 15 | d3bula2
|
|
 |
100.0 |
99 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 16 | c1xrsB_
|
|
 |
100.0 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:d-lysine 5,6-aminomutase beta subunit;
PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
|
| 17 | c1tx2A_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 18 | d1tx2a_
|
|
 |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 19 | d1j6ra_
|
|
 |
100.0 |
17 |
Fold:Methionine synthase activation domain-like
Superfamily:Methionine synthase activation domain-like
Family:Hypothetical protein TM0269 |
| 20 | d1ad1a_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 21 | d1ajza_ |
|
not modelled |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 22 | c2y5sA_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 23 | d1xrsb1 |
|
not modelled |
99.9 |
19 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 24 | c3tr9A_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 25 | c2vp8A_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
| 26 | d1eyea_ |
|
not modelled |
99.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 27 | c2dzaA_ |
|
not modelled |
99.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 28 | c1y80A_ |
|
not modelled |
99.9 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 29 | d3bula1 |
|
not modelled |
99.9 |
99 |
Fold:Methionine synthase domain-like
Superfamily:Methionine synthase domain
Family:Methionine synthase domain |
| 30 | c2h9aB_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 31 | c2vefB_ |
|
not modelled |
99.8 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 32 | d7reqa2 |
|
not modelled |
99.8 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 33 | d1ccwa_ |
|
not modelled |
99.8 |
22 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 34 | d1fmfa_ |
|
not modelled |
99.7 |
20 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 35 | c2yxbA_ |
|
not modelled |
99.6 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 36 | c2h9aA_ |
|
not modelled |
99.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 37 | c3mcnA_ |
|
not modelled |
99.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
PDBTitle: crystal structure of the 6-hyroxymethyl-7,8-dihydropterin2 pyrophosphokinase dihydropteroate synthase bifunctional enzyme from3 francisella tularensis
|
| 38 | c2bmbA_ |
|
not modelled |
99.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 39 | c3bicA_ |
|
not modelled |
99.1 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase, mitochondrial precursor;
PDBTitle: crystal structure of human methylmalonyl-coa mutase
|
| 40 | c3noyA_ |
|
not modelled |
99.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
| 41 | c3koxA_ |
|
not modelled |
98.4 |
29 |
PDB header:metal binding protein
Chain: A: PDB Molecule:d-ornithine aminomutase e component;
PDBTitle: crystal structure of ornithine 4,5 aminomutase in complex with 2,4-2 diaminobutyrate (anaerobic)
|
| 42 | c1e1cA_ |
|
not modelled |
97.8 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
| 43 | d1ka9f_ |
|
not modelled |
97.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 44 | d1xxxa1 |
|
not modelled |
96.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 45 | c2v9dB_ |
|
not modelled |
96.4 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 46 | c3s5oA_ |
|
not modelled |
96.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 47 | c3n2xB_ |
|
not modelled |
96.2 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 48 | c2ehhE_ |
|
not modelled |
96.1 |
13 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 49 | c3fkkA_ |
|
not modelled |
96.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
| 50 | c2r8wB_ |
|
not modelled |
95.9 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
| 51 | c3si9B_ |
|
not modelled |
95.8 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
| 52 | d1h5ya_ |
|
not modelled |
95.7 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 53 | c3e96B_ |
|
not modelled |
95.7 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 54 | d1o5ka_ |
|
not modelled |
95.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 55 | c3noeA_ |
|
not modelled |
95.6 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 56 | c3daqB_ |
|
not modelled |
95.6 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 57 | c2vc6A_ |
|
not modelled |
95.5 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 58 | c3eb2A_ |
|
not modelled |
95.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 59 | c2fdsA_ |
|
not modelled |
95.3 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:orotidine-monophosphate-decarboxylase;
PDBTitle: crystal structure of plasmodium berghei orotidine 5'-2 monophosphate decarboxylase (ortholog of plasmodium3 falciparum pf10_0225)
|
| 60 | d2fdsa1 |
|
not modelled |
95.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 61 | c3bi8A_ |
|
not modelled |
95.3 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 62 | d1xkya1 |
|
not modelled |
95.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 63 | c3na8A_ |
|
not modelled |
94.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 64 | c3g0sA_ |
|
not modelled |
94.8 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
| 65 | c3lerA_ |
|
not modelled |
94.8 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 66 | c2qw5B_ |
|
not modelled |
94.8 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase-like tim barrel;
PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
|
| 67 | d2a6na1 |
|
not modelled |
94.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 68 | c2qygC_ |
|
not modelled |
94.5 |
15 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
| 69 | c2oemA_ |
|
not modelled |
94.3 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate enolase;
PDBTitle: crystal structure of a rubisco-like protein from geobacillus2 kaustophilus liganded with mg2+ and 2,3-diketohexane 1-phosphate
|
| 70 | c3fluD_ |
|
not modelled |
94.2 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 71 | c6reqB_ |
|
not modelled |
94.2 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:protein (methylmalonyl-coa mutase);
PDBTitle: methylmalonyl-coa mutase, 3-carboxypropyl-coa inhibitor complex
|
| 72 | d1u1ha2 |
|
not modelled |
93.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Cobalamin-independent methionine synthase |
| 73 | c2nx9B_ |
|
not modelled |
93.7 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
| 74 | c3qfwB_ |
|
not modelled |
93.6 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase large
PDBTitle: crystal structure of rubisco-like protein from rhodopseudomonas2 palustris
|
| 75 | d2ffca1 |
|
not modelled |
93.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 76 | c3qfeB_ |
|
not modelled |
93.2 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
| 77 | c3cprB_ |
|
not modelled |
93.2 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 78 | c2yxgD_ |
|
not modelled |
92.9 |
10 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 79 | c2d69B_ |
|
not modelled |
92.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: crystal structure of the complex of sulfate ion and octameric2 ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from3 pyrococcus horikoshii ot3 (form-2 crystal)
|
| 80 | d1jvna1 |
|
not modelled |
92.7 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 81 | c2rfgB_ |
|
not modelled |
92.6 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 82 | d1vhna_ |
|
not modelled |
92.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 83 | c2y0fD_ |
|
not modelled |
92.1 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 84 | c1jvnB_ |
|
not modelled |
91.8 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 85 | c2y85D_ |
|
not modelled |
91.8 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:phosphoribosyl isomerase a;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
|
| 86 | c1telA_ |
|
not modelled |
91.7 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase, large subunit;
PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
|
| 87 | d1qh8b_ |
|
not modelled |
91.7 |
13 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 88 | c2r94B_ |
|
not modelled |
91.7 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 89 | d1ykwa1 |
|
not modelled |
91.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 90 | d2q8za1 |
|
not modelled |
91.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 91 | d1m1na_ |
|
not modelled |
91.0 |
14 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 92 | d1hl2a_ |
|
not modelled |
90.8 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 93 | d2d69a1 |
|
not modelled |
90.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 94 | c3obkH_ |
|
not modelled |
90.7 |
18 |
PDB header:lyase
Chain: H: PDB Molecule:delta-aminolevulinic acid dehydratase;
PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
|
| 95 | c3d0cB_ |
|
not modelled |
90.5 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
|
| 96 | d1thfd_ |
|
not modelled |
90.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 97 | c3dz1A_ |
|
not modelled |
90.1 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 98 | d1f74a_ |
|
not modelled |
89.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 99 | d1m1nb_ |
|
not modelled |
89.6 |
13 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 100 | d1rd5a_ |
|
not modelled |
89.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 101 | d7reqb2 |
|
not modelled |
89.3 |
17 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 102 | c3r89A_ |
|
not modelled |
89.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: crystal structure of orotidine 5-phosphate decarboxylase from2 anaerococcus prevotii dsm 20548
|
| 103 | c3eooL_ |
|
not modelled |
89.2 |
23 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 104 | d1muma_ |
|
not modelled |
89.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 105 | d8ruca1 |
|
not modelled |
89.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 106 | c3pueA_ |
|
not modelled |
89.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 107 | d1o66a_ |
|
not modelled |
88.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 108 | c3b0vD_ |
|
not modelled |
88.4 |
17 |
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
|
| 109 | c3qw3B_ |
|
not modelled |
87.8 |
16 |
PDB header:transferase, lyase
Chain: B: PDB Molecule:orotidine-5-phosphate decarboxylase/orotate
PDBTitle: structure of leishmania donovani omp decarboxylase
|
| 110 | c3nwrA_ |
|
not modelled |
87.3 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
| 111 | c3lciA_ |
|
not modelled |
86.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
| 112 | c1rldB_ |
|
not modelled |
86.5 |
21 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose 1,5 bisphosphate carboxylase/oxygenase (large
PDBTitle: solid-state phase transition in the crystal structure of ribulose 1,5-2 biphosphate carboxylase(slash)oxygenase
|
| 113 | d1wdda1 |
|
not modelled |
86.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 114 | c3lmaC_ |
|
not modelled |
85.9 |
19 |
PDB header:membrane protein
Chain: C: PDB Molecule:stage v sporulation protein ad (spovad);
PDBTitle: crystal structure of the stage v sporulation protein ad2 (spovad) from bacillus licheniformis. northeast structural3 genomics consortium target bir6.
|
| 115 | c3ru6C_ |
|
not modelled |
85.8 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: 1.8 angstrom resolution crystal structure of orotidine 5'-phosphate2 decarboxylase (pyrf) from campylobacter jejuni subsp. jejuni nctc3 11168
|
| 116 | d1vzwa1 |
|
not modelled |
85.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 117 | c1rcxH_ |
|
not modelled |
85.6 |
12 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
| 118 | c1ypxA_ |
|
not modelled |
85.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:putative vitamin-b12 independent methionine synthase family
PDBTitle: crystal structure of the putative vitamin-b12 independent methionine2 synthase from listeria monocytogenes, northeast structural genomics3 target lmr13
|
| 119 | c3lyeA_ |
|
not modelled |
84.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 120 | c2ftpA_ |
|
not modelled |
84.3 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|