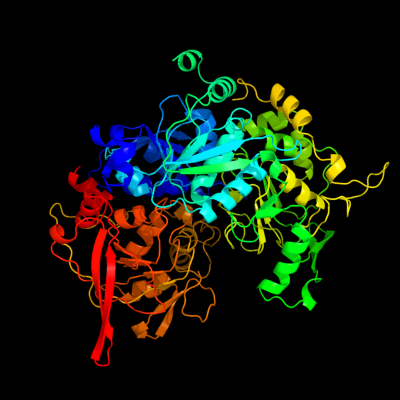
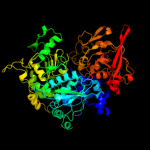
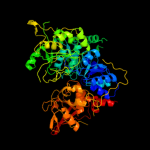
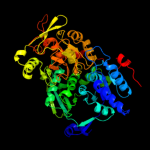
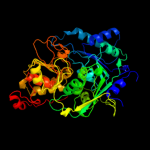
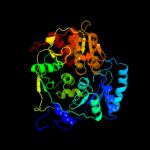
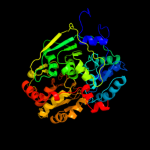
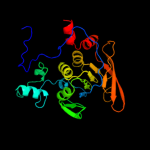
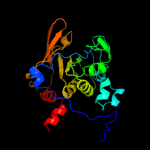
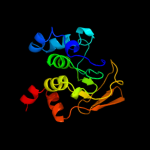
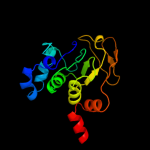
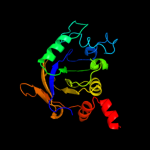
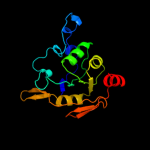
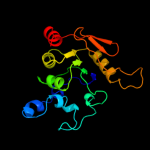
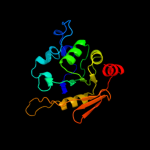
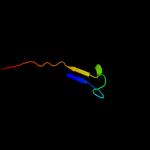
1 c2b3yB_
100.0
22
PDB header: lyaseChain: B: PDB Molecule: iron-responsive element binding protein 1;PDBTitle: structure of a monoclinic crystal form of human cytosolic aconitase2 (irp1)
2 c5acnA_
100.0
24
PDB header: lyase(carbon-oxygen)Chain: A: PDB Molecule: aconitase;PDBTitle: structure of activated aconitase. formation of the (4fe-4s)2 cluster in the crystal
3 d2b3ya2
100.0
23
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain4 d1acoa2
100.0
26
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain5 d1l5ja3
100.0
22
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain6 c1l5jB_
100.0
21
PDB header: lyaseChain: B: PDB Molecule: aconitate hydratase 2;PDBTitle: crystal structure of e. coli aconitase b.
7 d2b3ya1
100.0
19
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like8 d1acoa1
100.0
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like9 d1v7la_
100.0
24
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like10 c2pkpA_
100.0
28
PDB header: lyaseChain: A: PDB Molecule: homoaconitase small subunit;PDBTitle: crystal structure of 3-isopropylmalate dehydratase (leud)2 from methhanocaldococcus jannaschii dsm2661 (mj1271)
11 d1l5ja2
100.0
20
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like12 c2hcuA_
100.0
20
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus2 mutans
13 c3q3wB_
100.0
18
PDB header: transferaseChain: B: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: isopropylmalate isomerase small subunit from campylobacter jejuni.
14 c3h5jA_
100.0
18
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: leud_1-168 small subunit of isopropylmalate isomerase (rv2987c) from2 mycobacterium tuberculosis
15 d1g4ma1
86.9
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Arrestin/Vps26-like16 c1cjyB_
85.0
11
PDB header: hydrolaseChain: B: PDB Molecule: protein (cytosolic phospholipase a2);PDBTitle: human cytosolic phospholipase a2
17 d1cf1a1
80.3
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Arrestin/Vps26-like18 c2k6zA_
64.6
21
PDB header: metal transportChain: A: PDB Molecule: putative uncharacterized protein ttha1943;PDBTitle: solution structures of copper loaded form pcua (trans2 conformation of the peptide bond involving the nitrogen of3 p14)
19 d1x9la_
54.5
16
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: DR1885-like metal-binding proteinFamily: DR1885-like metal-binding protein20 c3dc1A_
50.5
12
PDB header: transferaseChain: A: PDB Molecule: kynurenine/alpha-aminoadipate aminotransferasePDBTitle: crystal structure of kynurenine aminotransferase ii complex with2 alpha-ketoglutarate
21 c2yx6C_
not modelled
42.3
10
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein ph0822;PDBTitle: crystal structure of ph0822
22 c3l4gI_
not modelled
38.0
19
PDB header: ligaseChain: I: PDB Molecule: phenylalanyl-trna synthetase alpha chain;PDBTitle: crystal structure of homo sapiens cytoplasmic phenylalanyl-trna2 synthetase
23 d1rdua_
not modelled
37.0
10
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like24 d1t3va_
not modelled
34.5
15
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like25 c3bolB_
not modelled
30.7
16
PDB header: transferaseChain: B: PDB Molecule: 5-methyltetrahydrofolate s-homocysteinePDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
26 d1o13a_
not modelled
30.5
11
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like27 d2r4qa1
not modelled
30.5
21
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like28 d2q7wa1
not modelled
30.4
8
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like29 d1yaaa_
not modelled
29.8
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like30 c3mo4B_
not modelled
29.0
20
PDB header: hydrolaseChain: B: PDB Molecule: alpha-1,3/4-fucosidase;PDBTitle: the crystal structure of an alpha-(1-3,4)-fucosidase from2 bifidobacterium longum subsp. infantis atcc 15697
31 c3gzaB_
not modelled
25.8
24
PDB header: hydrolaseChain: B: PDB Molecule: putative alpha-l-fucosidase;PDBTitle: crystal structure of putative alpha-l-fucosidase (np_812709.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.60 a resolution
32 c2yb1A_
not modelled
24.9
30
PDB header: hydrolaseChain: A: PDB Molecule: amidohydrolase;PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
33 d1t0tv_
not modelled
24.3
14
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Chlorite dismutase-like34 c2wbrA_
not modelled
23.6
31
PDB header: dna-binding proteinChain: A: PDB Molecule: gw182;PDBTitle: the rrm domain in gw182 proteins contributes to mirna-2 mediated gene silencing
35 c2wvsD_
not modelled
23.1
20
PDB header: hydrolaseChain: D: PDB Molecule: alpha-l-fucosidase;PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
36 d2csta_
not modelled
22.1
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like37 d1p90a_
not modelled
21.6
10
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: Nitrogenase accessory factor38 d3bofa2
not modelled
21.6
29
Fold: TIM beta/alpha-barrelSuperfamily: Homocysteine S-methyltransferaseFamily: Homocysteine S-methyltransferase39 c3qguB_
not modelled
20.8
22
PDB header: transferaseChain: B: PDB Molecule: ll-diaminopimelate aminotransferase;PDBTitle: l,l-diaminopimelate aminotransferase from chalmydomonas reinhardtii
40 d1vdha_
not modelled
20.6
9
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Chlorite dismutase-like41 d1go3e1
not modelled
20.5
12
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like42 c3ihjA_
not modelled
20.5
5
PDB header: transferaseChain: A: PDB Molecule: alanine aminotransferase 2;PDBTitle: human alanine aminotransferase 2 in complex with plp
43 d1eo1a_
not modelled
20.2
15
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like44 c3cmwA_
not modelled
19.9
19
PDB header: recombination/dnaChain: A: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
45 c3a2kB_
not modelled
19.9
8
PDB header: ligase/rnaChain: B: PDB Molecule: trna(ile)-lysidine synthase;PDBTitle: crystal structure of tils complexed with trna
46 c1pt1B_
not modelled
19.8
16
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
47 d1ppya_
not modelled
19.8
16
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC48 c2wfbA_
not modelled
18.8
16
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein orp;PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
49 d2ay1a_
not modelled
18.1
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like50 c3pg8B_
not modelled
17.2
11
PDB header: transferaseChain: B: PDB Molecule: phospho-2-dehydro-3-deoxyheptonate aldolase;PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
51 d7aata_
not modelled
16.8
11
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like52 c3eypB_
not modelled
16.5
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative alpha-l-fucosidase;PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
53 c3gitA_
not modelled
16.4
32
PDB header: transferaseChain: A: PDB Molecule: carbon monoxide dehydrogenase/acetyl-coa synthase subunitPDBTitle: crystal structure of a truncated acetyl-coa synthase
54 d1lt7a_
not modelled
16.1
25
Fold: TIM beta/alpha-barrelSuperfamily: Homocysteine S-methyltransferaseFamily: Homocysteine S-methyltransferase55 c2nvgA_
not modelled
15.9
14
PDB header: oxidoreductaseChain: A: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: soluble domain of rieske iron sulfur protein.
56 d3cdda2
not modelled
15.5
15
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like57 c2vxhF_
not modelled
15.3
0
PDB header: oxidoreductaseChain: F: PDB Molecule: chlorite dismutase;PDBTitle: the crystal structure of chlorite dismutase: a detox enzyme2 producing molecular oxygen
58 c3k7yA_
not modelled
14.8
10
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: aspartate aminotransferase of plasmodium falciparum
59 d1x0ma1
not modelled
14.5
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like60 d1w7la_
not modelled
14.0
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like61 c2c45F_
not modelled
13.9
11
PDB header: lyaseChain: F: PDB Molecule: aspartate 1-decarboxylase precursor;PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
62 c3c9uB_
not modelled
13.4
13
PDB header: transferaseChain: B: PDB Molecule: thiamine monophosphate kinase;PDBTitle: aathil complexed with adp and tpp
63 c2ebbA_
not modelled
13.0
15
PDB header: lyaseChain: A: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine2 dehydratase (pterin carbinolamine dehydratase) from3 geobacillus kaustophilus hta426
64 c3pplB_
not modelled
12.7
14
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of an aspartate transaminase (ncgl0237, cgl0240)2 from corynebacterium glutamicum atcc 13032 kitasato at 1.25 a3 resolution
65 d2r5ea1
not modelled
12.6
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like66 c3nn4C_
not modelled
12.5
5
PDB header: oxidoreductaseChain: C: PDB Molecule: chlorite dismutase;PDBTitle: structure of chlorite dismutase from candidatus nitrospira defluvii2 r173k mutant
67 c2oxlA_
not modelled
12.3
21
PDB header: gene regulationChain: A: PDB Molecule: hypothetical protein ymgb;PDBTitle: structure and function of the e. coli protein ymgb: a protein critical2 for biofilm formation and acid resistance
68 d1l0sa_
not modelled
12.2
21
Fold: Single-stranded left-handed beta-helixSuperfamily: An insect antifreeze proteinFamily: An insect antifreeze protein69 c3m84A_
not modelled
12.1
13
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylformylglycinamidine cyclo-ligase;PDBTitle: crystal structure of phosphoribosylaminoimidazole synthetase from2 francisella tularensis
70 c2jz2A_
not modelled
12.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ssl0352 protein;PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
71 c3muxB_
not modelled
12.0
13
PDB header: lyaseChain: B: PDB Molecule: putative 4-hydroxy-2-oxoglutarate aldolase;PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
72 d2nu7b2
not modelled
11.9
25
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Succinyl-CoA synthetase, beta-chain, N-terminal domain73 c3m6yA_
not modelled
11.7
13
PDB header: lyaseChain: A: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase;PDBTitle: structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at2 1.45 a resolution.
74 d1w44a_
not modelled
11.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)75 c1ni5A_
not modelled
11.4
7
PDB header: cell cycleChain: A: PDB Molecule: putative cell cycle protein mesj;PDBTitle: structure of the mesj pp-atpase from escherichia coli
76 c2w9mB_
not modelled
11.3
25
PDB header: dna replicationChain: B: PDB Molecule: polymerase x;PDBTitle: structure of family x dna polymerase from deinococcus2 radiodurans
77 c3kxyW_
not modelled
10.7
25
PDB header: chaperone/transcription inhibitorChain: W: PDB Molecule: exse;PDBTitle: crystal structure of the exsc-exse complex
78 c3cmuA_
not modelled
10.7
16
PDB header: recombination/dnaChain: A: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
79 d1m8na_
not modelled
10.3
14
Fold: Single-stranded left-handed beta-helixSuperfamily: An insect antifreeze proteinFamily: An insect antifreeze protein80 d2p5zx2
not modelled
10.2
25
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like81 d1ru3a_
not modelled
10.1
23
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Acetyl-CoA synthase82 c3eibB_
not modelled
9.8
19
PDB header: transferaseChain: B: PDB Molecule: ll-diaminopimelate aminotransferase;PDBTitle: crystal structure of k270n variant of ll-diaminopimelate2 aminotransferase from arabidopsis thaliana
83 c2gp4A_
not modelled
9.8
24
PDB header: lyaseChain: A: PDB Molecule: 6-phosphogluconate dehydratase;PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
84 d1oaoc_
not modelled
9.7
32
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Acetyl-CoA synthase85 c3bzwA_
not modelled
9.6
12
PDB header: hydrolaseChain: A: PDB Molecule: putative lipase;PDBTitle: crystal structure of a putative lipase from bacteroides2 thetaiotaomicron
86 d3bzwa1
not modelled
9.6
12
Fold: Flavodoxin-likeSuperfamily: SGNH hydrolaseFamily: BT2961-like87 c3bkdB_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: B: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
88 c3bkdG_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: G: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
89 c3bkdD_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: D: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
90 c3bkdC_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: C: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
91 c3bkdE_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: E: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
92 c3bkdF_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: F: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
93 c3bkdA_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: A: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
94 c3bkdH_
not modelled
9.6
38
PDB header: viral protein, membrane proteinChain: H: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
95 d1qlma_
not modelled
9.4
15
Fold: Methenyltetrahydromethanopterin cyclohydrolaseSuperfamily: Methenyltetrahydromethanopterin cyclohydrolaseFamily: Methenyltetrahydromethanopterin cyclohydrolase96 c2kadC_
not modelled
9.4
38
PDB header: membrane proteinChain: C: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
97 c2kadA_
not modelled
9.4
38
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
98 c2kadB_
not modelled
9.4
38
PDB header: membrane proteinChain: B: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
99 c2kadD_
not modelled
9.4
38
PDB header: membrane proteinChain: D: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain