| 1 | c3klzE_ |
|
 |
100.0 |
55 |
PDB header:membrane protein
Chain: E: PDB Molecule:putative formate transporter 1;
PDBTitle: pentameric formate channel with formate bound
|
| 2 | c3kcvG_ |
|
 |
100.0 |
100 |
PDB header:transport protein
Chain: G: PDB Molecule:probable formate transporter 1;
PDBTitle: structure of formate channel
|
| 3 | c1ldaA_ |
|
 |
95.6 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:glycerol uptake facilitator protein;
PDBTitle: crystal structure of the e. coli glycerol facilitator (glpf) without2 substrate glycerol
|
| 4 | d1fx8a_ |
|
 |
95.6 |
14 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 5 | c3llqB_ |
|
 |
93.6 |
11 |
PDB header:membrane protein
Chain: B: PDB Molecule:aquaporin z 2;
PDBTitle: aquaporin structure from plant pathogen agrobacterium tumerfaciens
|
| 6 | c2b5fD_ |
|
 |
91.0 |
11 |
PDB header:transport protein,membrane protein
Chain: D: PDB Molecule:aquaporin;
PDBTitle: crystal structure of the spinach aquaporin sopip2;1 in an2 open conformation to 3.9 resolution
|
| 7 | d1rc2a_ |
|
 |
88.9 |
11 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 8 | c3iyzA_ |
|
 |
88.4 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:aquaporin-4;
PDBTitle: structure of aquaporin-4 s180d mutant at 10.0 a resolution from2 electron micrograph
|
| 9 | c3gd8A_ |
|
 |
84.6 |
17 |
PDB header:membrane protein
Chain: A: PDB Molecule:aquaporin-4;
PDBTitle: crystal structure of human aquaporin 4 at 1.8 and its mechanism of2 conductance
|
| 10 | c2w2eA_ |
|
 |
79.5 |
12 |
PDB header:membrane protein
Chain: A: PDB Molecule:aquaporin;
PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin,2 aqy1, in a closed conformation at ph 3.5
|
| 11 | d1j4na_ |
|
 |
77.9 |
12 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 12 | c2d57A_ |
|
 |
77.6 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:aquaporin-4;
PDBTitle: double layered 2d crystal structure of aquaporin-4 (aqp4m23) at 3.2 a2 resolution by electron crystallography
|
| 13 | c3c02A_ |
|
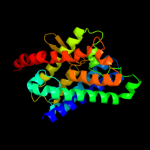 |
73.3 |
9 |
PDB header:membrane protein
Chain: A: PDB Molecule:aquaglyceroporin;
PDBTitle: x-ray structure of the aquaglyceroporin from plasmodium falciparum
|
| 14 | c3d9sB_ |
|
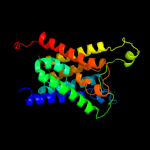 |
50.3 |
12 |
PDB header:membrane protein
Chain: B: PDB Molecule:aquaporin-5;
PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
|
| 15 | c2f2bA_ |
|
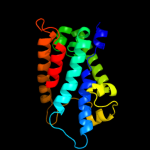 |
48.5 |
9 |
PDB header:membrane protein
Chain: A: PDB Molecule:aquaporin aqpm;
PDBTitle: crystal structure of integral membrane protein aquaporin aqpm at 1.68a2 resolution
|
| 16 | d1w2za2 |
|
 |
46.4 |
25 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 17 | c1ymgA_ |
|
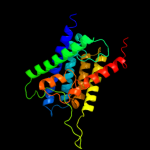 |
46.0 |
14 |
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
|
| 18 | d1ymga1 |
|
 |
46.0 |
14 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 19 | d1d6za2 |
|
 |
41.9 |
21 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 20 | d2oqea2 |
|
 |
41.3 |
25 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 21 | d1w6ga2 |
|
not modelled |
40.6 |
25 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 22 | d1h6ia_ |
|
not modelled |
21.0 |
9 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 23 | c3e49A_ |
|
not modelled |
16.1 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849 with a tim barrel fold;
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
|
| 24 | c3lotC_ |
|
not modelled |
15.5 |
18 |
PDB header:structure genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
|
| 25 | c3c6cA_ |
|
not modelled |
15.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
|
| 26 | c2y7eA_ |
|
not modelled |
13.6 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
|
| 27 | d1c17m_ |
|
not modelled |
13.6 |
31 |
Fold:F1F0 ATP synthase subunit A
Superfamily:F1F0 ATP synthase subunit A
Family:F1F0 ATP synthase subunit A |
| 28 | c1ksiA_ |
|
not modelled |
12.9 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure of a eukaryotic (pea seedling) copper-containing2 amine oxidase at 2.2a resolution
|
| 29 | c3loyB_ |
|
not modelled |
12.5 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure of a copper-containing benzylamine oxidase from2 hansenula polymorpha
|
| 30 | c3no5C_ |
|
not modelled |
12.1 |
20 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
|
| 31 | c3chvA_ |
|
not modelled |
11.9 |
28 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
| 32 | c1d6uB_ |
|
not modelled |
11.7 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure of e. coli amine oxidase anaerobically reduced with2 beta-phenylethylamine
|
| 33 | c1ui7A_ |
|
not modelled |
11.5 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phenylethylamine oxidase;
PDBTitle: site-directed mutagenesis of his433 involved in binding of2 copper ion in arthrobacter globiformis amine oxidase
|
| 34 | c1ekmC_ |
|
not modelled |
10.5 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure at 2.5 a resolution of zinc-substituted2 copper amine oxidase of hansenula polymorpha expressed in3 escherichia coli
|
| 35 | c3nbbC_ |
|
not modelled |
10.1 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:peroxisomal primary amine oxidase;
PDBTitle: crystal structure of mutant y305f expressed in e. coli in the copper2 amine oxidase from hansenula polymorpha
|
| 36 | c3e02A_ |
|
not modelled |
9.3 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
| 37 | c2jy0A_ |
|
not modelled |
8.5 |
50 |
PDB header:membrane protein, viral protein
Chain: A: PDB Molecule:protease ns2-3;
PDBTitle: solution nmr structure of hcv ns2 protein, membrane segment2 (1-27)
|
| 38 | d2c1wa1 |
|
not modelled |
8.2 |
67 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Eukaryotic EndoU ribonuclease |
| 39 | d1rm6b2 |
|
not modelled |
7.7 |
25 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 40 | d2foka2 |
|
not modelled |
7.0 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Restriction endonuclease FokI, N-terminal (recognition) domain |
| 41 | c1rm6E_ |
|
not modelled |
6.9 |
25 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:4-hydroxybenzoyl-coa reductase beta subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
| 42 | c2voyG_ |
|
not modelled |
5.5 |
39 |
PDB header:hydrolase
Chain: G: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
|

