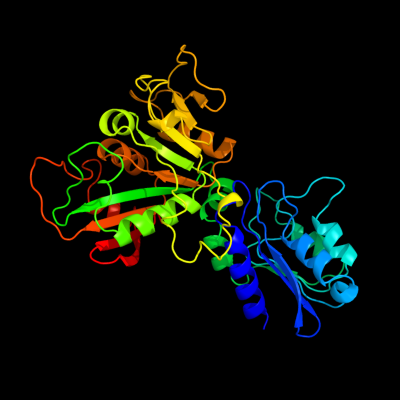
1 c2d5nB_
100.0
42
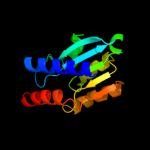
PDB header: hydrolase, oxidoreductaseChain: B: PDB Molecule: riboflavin biosynthesis protein ribd;PDBTitle: crystal structure of a bifunctional deaminase and reductase2 involved in riboflavin biosynthesis
2 c2o7pA_
100.0
97
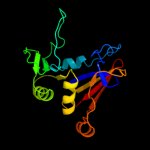
PDB header: hydrolase, oxidoreductaseChain: A: PDB Molecule: riboflavin biosynthesis protein ribd;PDBTitle: the crystal structure of ribd from escherichia coli in complex with2 the oxidised nadp+ cofactor in the active site of the reductase3 domain
3 c2hxvA_
100.0
34
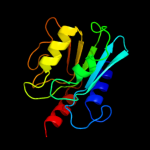
PDB header: biosynthetic proteinChain: A: PDB Molecule: diaminohydroxyphosphoribosylaminopyrimidine deaminase/ 5-PDBTitle: crystal structure of a diaminohydroxyphosphoribosylaminopyrimidine2 deaminase/ 5-amino-6-(5-phosphoribosylamino)uracil reductase (tm1828)3 from thermotoga maritima at 1.80 a resolution
4 d2b3za1
100.0
32
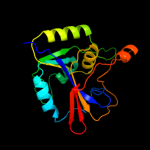
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: RibD C-terminal domain-like5 d2azna1
100.0
32
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: RibD C-terminal domain-like6 c2p4gA_
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a pyrimidine reductase-like protein (dip1392)2 from corynebacterium diphtheriae nctc at 2.30 a resolution
7 d2b3za2
100.0
57
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like8 d2hxva1
100.0
26
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: RibD C-terminal domain-like9 d2hxva2
100.0
44
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like10 d1z3aa1
100.0
33
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like11 c3ocqA_
100.0
33
PDB header: hydrolaseChain: A: PDB Molecule: putative cytosine/adenosine deaminase;PDBTitle: crystal structure of trna-specific adenosine deaminase from salmonella2 enterica
12 c2nx8A_
100.0
28
PDB header: hydrolaseChain: A: PDB Molecule: trna-specific adenosine deaminase;PDBTitle: the crystal structure of the trna-specific adenosine deaminase from2 streptococcus pyogenes
13 d2b3ja1
100.0
30
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like14 d2g84a1
100.0
25
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like15 c3kgyA_
100.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: bifunctional deaminase-reductase domain protein;PDBTitle: crystal structure of putative dihydrofolate reductase (yp_001636057.1)2 from chloroflexus aurantiacus j-10-fl at 1.50 a resolution
16 d1wwra1
100.0
30
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like17 d1p6oa_
100.0
28
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like18 c3jtwB_
100.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: dihydrofolate reductase;PDBTitle: crystal structure of putative dihydrofolate reductase (yp_805003.1)2 from pediococcus pentosaceus atcc 25745 at 1.90 a resolution
19 c3dh1D_
100.0
29
PDB header: hydrolaseChain: D: PDB Molecule: trna-specific adenosine deaminase 2;PDBTitle: crystal structure of human trna-specific adenosine-34 deaminase2 subunit adat2
20 c2gd9A_
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein yyap;PDBTitle: crystal structure of a putative dihydrofolate reductase (bsu40760,2 yyap) from bacillus subtilis at 2.30 a resolution
21 c2xw7A_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrofolate reductase;PDBTitle: structure of mycobacterium smegmatis putative reductase ms0308
22 d1d1ga_
not modelled
100.0
21
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases23 d1wkqa_
not modelled
100.0
27
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like24 d1vq2a_
not modelled
100.0
36
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like25 d2a8na1
not modelled
100.0
33
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like26 c2w4lC_
not modelled
100.0
32
PDB header: hydrolaseChain: C: PDB Molecule: deoxycytidylate deaminase;PDBTitle: human dcmp deaminase
27 c2hvwC_
not modelled
100.0
31
PDB header: hydrolaseChain: C: PDB Molecule: deoxycytidylate deaminase;PDBTitle: crystal structure of dcmp deaminase from streptococcus2 mutans
28 c3ky8B_
not modelled
100.0
21
PDB header: biosynthetic proteinChain: B: PDB Molecule: putative riboflavin biosynthesis protein;PDBTitle: crystal structure of putative riboflavin biosynthesis protein2 (yp_001092907.1) from shewanella sp. pv-4 at 2.12 a resolution
29 d1seja1
not modelled
99.7
14
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases30 d1vdra_
not modelled
99.6
13
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases31 c3ix9B_
not modelled
99.5
18
PDB header: oxidoreductaseChain: B: PDB Molecule: dihydrofolate reductase;PDBTitle: crystal structure of streptococcus pneumoniae dihydrofolate2 reductase - sp9 mutant
32 d3dfra_
not modelled
99.4
15
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases33 d1kmva_
not modelled
99.4
16
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases34 c3k2hA_
not modelled
99.3
17
PDB header: transferaseChain: A: PDB Molecule: dihydrofolate reductase/thymidylate synthase;PDBTitle: co-crystal structure of dihydrofolate reductase/thymidylate synthase2 from babesia bovis with dump, pemetrexed and nadp
35 d8dfra_
not modelled
99.3
18
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases36 d2fzia1
not modelled
99.2
20
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases37 c3e0bA_
not modelled
99.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrofolate reductase;PDBTitle: bacillus anthracis dihydrofolate reductase complexed with2 nadph and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-3 ynyl)-6-ethylpyrimidine (ucp120b)
38 c1zdrB_
not modelled
99.2
18
PDB header: oxidoreductaseChain: B: PDB Molecule: dihydrofolate reductase;PDBTitle: dhfr from bacillus stearothermophilus
39 d1df7a_
not modelled
99.2
17
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases40 c3tq8A_
not modelled
99.2
17
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: dihydrofolate reductase;PDBTitle: structure of the dihydrofolate reductase (fola) from coxiella burnetii2 in complex with trimethoprim
41 c3ia5A_
not modelled
99.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrofolate reductase;PDBTitle: moritella profunda dihydrofolate reductase (dhfr)
42 d1ra9a_
not modelled
99.1
14
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases43 c2oipE_
not modelled
99.0
18
PDB header: transferase, oxidoreductaseChain: E: PDB Molecule: chain a, crystal structure of dhfr;PDBTitle: crystal structure of the s290g active site mutant of ts-2 dhfr from cryptosporidium hominis
44 c3f0uX_
not modelled
99.0
16
PDB header: oxidoreductaseChain: X: PDB Molecule: trimethoprim-sensitive dihydrofolate reductase;PDBTitle: staphylococcus aureus f98y mutant dihydrofolate reductase2 complexed with nadph and 2,4-diamino-5-[3-(3-methoxy-5-3 phenylphenyl)but-1-ynyl]-6-methylpyrimidine
45 d1j3ka_
not modelled
98.9
12
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases46 d1aoea_
not modelled
98.8
12
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases47 c2blcA_
not modelled
98.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrofolate reductase-thymidylate synthase;PDBTitle: sp21 double mutant p. vivax dihydrofolate reductase in2 complex with des-chloropyrimethamine
48 c3dg8B_
not modelled
98.7
13
PDB header: oxidoreductase, transferaseChain: B: PDB Molecule: bifunctional dihydrofolate reductase-thymidylatePDBTitle: quadruple mutant (n51i+c59r+s108n+i164l) plasmodium2 falciparum dihydrofolate reductase-thymidylate synthase3 (pfdhfr-ts) complexed with rjf670, nadph, and dump
49 c3jsuA_
not modelled
98.7
12
PDB header: oxidoreductase, transferaseChain: A: PDB Molecule: dihydrofolate reductase-thymidylate synthase;PDBTitle: quadruple mutant(n51i+c59r+s108n+i164l) plasmodium falciparum2 dihydrofolate reductase-thymidylate synthase(pfdhfr-ts) complexed3 with qn254, nadph, and dump
50 c3clbA_
not modelled
98.7
14
PDB header: oxidoreductase, transferaseChain: A: PDB Molecule: dhfr-ts;PDBTitle: structure of bifunctional tcdhfr-ts in complex with tmq
51 c3cseA_
not modelled
98.5
15
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrofolate reductase;PDBTitle: candida glabrata dihydrofolate reductase complexed with2 nadph and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-3 ynyl)-6-ethylpyrimidine (ucp120b)
52 c3rg9A_
not modelled
98.1
16
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: bifunctional dihydrofolate reductase-thymidylate synthase;PDBTitle: trypanosoma brucei dihydrofolate reductase (tbdhfr) in complex with2 wr99210
53 d1uwza_
not modelled
97.7
21
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase54 c3g8qA_
not modelled
97.7
34
PDB header: rna binding proteinChain: A: PDB Molecule: predicted rna-binding protein, contains thumpPDBTitle: a cytidine deaminase edits c-to-u in transfer rnas in2 archaea
55 d2d30a1
not modelled
97.6
21
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase56 d1juva_
not modelled
97.5
22
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases57 d1alna1
not modelled
97.4
26
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase58 c3ijfX_
not modelled
97.3
19
PDB header: hydrolaseChain: X: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase from mycobacterium2 tuberculosis
59 d1r5ta_
not modelled
97.2
20
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase60 c3dmoD_
not modelled
97.1
25
PDB header: hydrolaseChain: D: PDB Molecule: cytidine deaminase;PDBTitle: 1.6 a crystal structure of cytidine deaminase from2 burkholderia pseudomallei
61 c1alnA_
not modelled
97.0
25
PDB header: hydrolaseChain: A: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase complexed with 3-deazacytidine
62 d1mq0a_
not modelled
97.0
20
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase63 c3r2nC_
not modelled
96.9
19
PDB header: hydrolaseChain: C: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase from mycobacterium leprae
64 c2nytB_
not modelled
96.8
19
PDB header: hydrolaseChain: B: PDB Molecule: probable c->u-editing enzyme apobec-2;PDBTitle: the apobec2 crystal structure and functional implications2 for aid
65 d2fr5a1
not modelled
96.8
14
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase66 c3oj6C_
not modelled
96.3
24
PDB header: hydrolaseChain: C: PDB Molecule: blasticidin-s deaminase;PDBTitle: crystal structure of blasticidin s deaminase from coccidioides immitis
67 d2z3ga1
not modelled
96.2
21
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase68 c3b8fB_
not modelled
96.1
14
PDB header: hydrolaseChain: B: PDB Molecule: putative blasticidin s deaminase;PDBTitle: crystal structure of the cytidine deaminase from bacillus anthracis
69 c2kboA_
not modelled
95.8
22
PDB header: hydrolaseChain: A: PDB Molecule: dna dc->du-editing enzyme apobec-3g;PDBTitle: structure, interaction, and real-time monitoring of the2 enzymatic reaction of wild type apobec3g
70 d1alna2
not modelled
95.7
16
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase71 d1j5ta_
not modelled
74.9
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes72 d1o60a_
not modelled
71.5
14
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase73 c3qjaA_
not modelled
66.4
15
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
74 d1zcza2
not modelled
65.3
42
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC75 c4a1oB_
not modelled
59.6
37
PDB header: transferase-hydrolaseChain: B: PDB Molecule: bifunctional purine biosynthesis protein purh;PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
76 c2h6rG_
not modelled
54.2
11
PDB header: isomeraseChain: G: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
77 c2gm2A_
not modelled
53.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
78 c2xecD_
not modelled
49.8
20
PDB header: isomeraseChain: D: PDB Molecule: putative maleate isomerase;PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
79 c1zczA_
not modelled
49.0
42
PDB header: transferase/hydrolaseChain: A: PDB Molecule: bifunctional purine biosynthesis protein purh;PDBTitle: crystal structure of phosphoribosylaminoimidazolecarboxamide2 formyltransferase / imp cyclohydrolase (tm1249) from thermotoga3 maritima at 1.88 a resolution
80 d1o13a_
not modelled
47.2
15
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like81 d1d9ea_
not modelled
46.8
14
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase82 c2wfbA_
not modelled
46.7
27
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein orp;PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
83 c1piiA_
not modelled
46.4
25
PDB header: bifunctional(isomerase and synthase)Chain: A: PDB Molecule: n-(5'phosphoribosyl)anthranilate isomerase;PDBTitle: three-dimensional structure of the bifunctional enzyme2 phosphoribosylanthranilate isomerase:3 indoleglycerolphosphate synthase from escherichia coli4 refined at 2.0 angstroms resolution
84 d1i4na_
not modelled
45.0
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes85 c2c3zA_
not modelled
44.1
14
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
86 d1piia2
not modelled
43.7
22
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes87 c2yx6C_
not modelled
40.7
16
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein ph0822;PDBTitle: crystal structure of ph0822
88 c2c4kD_
not modelled
40.1
8
PDB header: regulatory proteinChain: D: PDB Molecule: phosphoribosyl pyrophosphate synthetase-PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
89 c3sz8D_
not modelled
39.7
14
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 2;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
90 d1a53a_
not modelled
38.2
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes91 d2fb5a1
not modelled
37.7
35
Fold: YojJ-likeSuperfamily: YojJ-likeFamily: YojJ-like92 c2k2eA_
not modelled
37.3
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bp2786;PDBTitle: solution nmr structure of bordetella pertussis protein2 bp2786, a mth938-like domain. northeast structural3 genomics consortium target ber31
93 c2gewA_
not modelled
36.7
24
PDB header: oxidoreductaseChain: A: PDB Molecule: cholesterol oxidase;PDBTitle: atomic resolution structure of cholesterol oxidase @ ph 9.02 (streptomyces sp. sa-coo)
94 d2q4qa1
not modelled
36.5
21
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like95 d1xk7a1
not modelled
35.9
15
Fold: CoA-transferase family III (CaiB/BaiF)Superfamily: CoA-transferase family III (CaiB/BaiF)Family: CoA-transferase family III (CaiB/BaiF)96 d1g5ha1
not modelled
34.3
23
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS97 d1zbfa1
not modelled
33.0
23
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H98 c3gxgA_
not modelled
31.8
18
PDB header: hydrolaseChain: A: PDB Molecule: putative phosphatase (duf442);PDBTitle: crystal structure of putative phosphatase (duf442) (yp_001181608.1)2 from shewanella putrefaciens cn-32 at 1.60 a resolution
99 d1u9ya2
not modelled
31.1
23
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like100 d1vc4a_
not modelled
30.6
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes101 c3efhB_
not modelled
29.6
28
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase 1;PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
102 c3b0vD_
not modelled
29.4
11
PDB header: oxidoreductase/rnaChain: D: PDB Molecule: trna-dihydrouridine synthase;PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
103 d2f5va1
not modelled
29.3
24
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain104 d1ewna_
not modelled
28.7
28
Fold: FMT C-terminal domain-likeSuperfamily: FMT C-terminal domain-likeFamily: 3-methyladenine DNA glycosylase (AAG, ANPG, MPG)105 d1eo1a_
not modelled
28.7
23
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like106 c1u9yD_
not modelled
28.7
25
PDB header: transferaseChain: D: PDB Molecule: ribose-phosphate pyrophosphokinase;PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
107 d2fi9a1
not modelled
28.2
6
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like108 c3ggmB_
not modelled
27.7
30
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein bt9727_2919;PDBTitle: crystal structure of bt9727_2919 from bacillus2 thuringiensis subsp. northeast structural genomics target3 bur228b
109 c2jugB_
not modelled
27.5
21
PDB header: biosynthetic proteinChain: B: PDB Molecule: tubc protein;PDBTitle: multienzyme docking in hybrid megasynthetases
110 d1znna1
not modelled
26.8
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: PdxS-like111 d1rdua_
not modelled
26.5
23
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like112 d1dkua2
not modelled
26.5
25
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like113 d1wu7a1
not modelled
26.1
15
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS114 c1znnF_
not modelled
25.7
16
PDB header: biosynthetic proteinChain: F: PDB Molecule: plp synthase;PDBTitle: structure of the synthase subunit of plp synthase
115 d2g4ca1
not modelled
25.4
20
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS116 d1t3va_
not modelled
24.4
13
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like117 d1w0ma_
not modelled
24.0
32
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)118 d2vjma1
not modelled
23.5
9
Fold: CoA-transferase family III (CaiB/BaiF)Superfamily: CoA-transferase family III (CaiB/BaiF)Family: CoA-transferase family III (CaiB/BaiF)119 d2fvta1
not modelled
22.4
5
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like120 c2qtdA_
not modelled
22.3
8
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized protein mj0327;PDBTitle: crystal structure of a putative dinitrogenase (mj0327) from2 methanocaldococcus jannaschii dsm at 1.70 a resolution