1 c2zvyB_
100.0
94
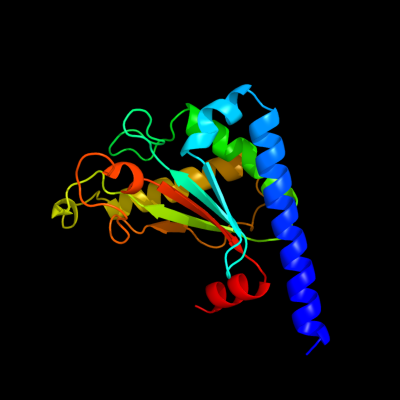
PDB header: membrane proteinChain: B: PDB Molecule: chemotaxis protein motb;PDBTitle: structure of the periplasmic domain of motb from salmonella2 (crystal form ii)
2 c3khnB_
100.0
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: motb protein, putative;PDBTitle: crystal structure of putative motb like protein dvu_22282 from desulfovibrio vulgaris.
3 c2zovA_
100.0
91
PDB header: membrane proteinChain: A: PDB Molecule: chemotaxis protein motb;PDBTitle: structure of the periplasmic domain of motb from salmonella2 (crystal form i)
4 c2l26A_
100.0
27
PDB header: membrane proteinChain: A: PDB Molecule: uncharacterized protein rv0899/mt0922;PDBTitle: rv0899 from mycobacterium tuberculosis contains two separated domains
5 c3cyqM_
100.0
21
PDB header: membrane proteinChain: M: PDB Molecule: chemotaxis protein motb;PDBTitle: the crystal structure of the complex of the c-terminal domain of2 helicobacter pylori motb (residues 125-256) with n-acetylmuramic acid
6 c2k1sA_
100.0
21
PDB header: lipoproteinChain: A: PDB Molecule: inner membrane lipoprotein yiad;PDBTitle: solution nmr structure of the folded c-terminal fragment of yiad from2 escherichia coli. northeast structural genomics consortium target3 er553.
7 c3ldtA_
99.9
19
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein, ompa family protein;PDBTitle: crystal structure of an outer membrane protein(ompa)from2 legionella pneumophila
8 c2kgwA_
99.9
26
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: solution structure of the carboxy-terminal domain of ompatb, a pore2 forming protein from mycobacterium tuberculosis
9 c3td4D_
99.9
25
PDB header: membrane protein,peptide binding proteinChain: D: PDB Molecule: outer membrane protein omp38;PDBTitle: crystal structure of ompa-like domain from acinetobacter baumannii in2 complex with diaminopimelate
10 d2aizp1
99.9
29
Fold: Bacillus chorismate mutase-likeSuperfamily: OmpA-likeFamily: OmpA-like11 d2hqsc1
99.9
24
Fold: Bacillus chorismate mutase-likeSuperfamily: OmpA-likeFamily: OmpA-like12 d1r1ma_
99.9
21
Fold: Bacillus chorismate mutase-likeSuperfamily: OmpA-likeFamily: OmpA-like13 c1r1mA_
99.9
21
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein class 4;PDBTitle: structure of the ompa-like domain of rmpm from neisseria2 meningitidis
14 c3oonA_
99.9
24
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein (tpn50);PDBTitle: the structure of an outer membrance protein from borrelia burgdorferi2 b31
15 c2zf8A_
99.5
18
PDB header: structural proteinChain: A: PDB Molecule: component of sodium-driven polar flagellar motor;PDBTitle: crystal structure of moty
16 c3l7pA_
58.4
13
PDB header: transcriptionChain: A: PDB Molecule: putative nitrogen regulatory protein pii;PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
17 d2cz4a1
42.2
13
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein18 c2ccaA_
34.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxidase/catalase t;PDBTitle: crystal structure of the catalase-peroxidase (katg) and2 s315t mutant from mycobacterium tuberculosis
19 c2rd5D_
29.4
21
PDB header: protein bindingChain: D: PDB Molecule: pii protein;PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
20 d1qy7a_
28.8
18
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein21 c2b2qB_
not modelled
26.8
23
PDB header: oxidoreductaseChain: B: PDB Molecule: catalase-peroxidase;PDBTitle: crystal structure of native catalase-peroxidase katg at2 ph7.5
22 c3mhyC_
not modelled
26.0
18
PDB header: signaling proteinChain: C: PDB Molecule: pii-like protein pz;PDBTitle: a new pii protein structure
23 c2fxhB_
not modelled
25.7
23
PDB header: oxidoreductaseChain: B: PDB Molecule: catalase-peroxidase protein;PDBTitle: crystal structure of katg at ph 6.5
24 c1itkB_
not modelled
25.4
21
PDB header: oxidoreductaseChain: B: PDB Molecule: catalase-peroxidase;PDBTitle: crystal structure of catalase-peroxidase from haloarcula2 marismortui
25 c2j9dG_
not modelled
24.8
20
PDB header: membrane transportChain: G: PDB Molecule: hypothetical nitrogen regulatory pii-likePDBTitle: structure of glnk1 with bound effectors indicates2 regulatory mechanism for ammonia uptake
26 d2ns1b1
not modelled
23.9
18
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein27 d2h8pc1
not modelled
22.5
13
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels28 d2piia_
not modelled
22.0
14
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein29 c1skhA_
not modelled
21.7
30
PDB header: unknown functionChain: A: PDB Molecule: major prion protein 2;PDBTitle: n-terminal (1-30) of bovine prion protein
30 d2axla1
not modelled
21.7
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RecQ helicase DNA-binding domain-like31 d1saza1
not modelled
17.3
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like32 c3o8wA_
not modelled
17.1
11
PDB header: signaling proteinChain: A: PDB Molecule: nitrogen regulatory protein p-ii (glnb-1);PDBTitle: archaeoglobus fulgidus glnk1
33 d1ofda2
not modelled
17.1
34
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases34 c1ub2A_
not modelled
14.7
33
PDB header: oxidoreductaseChain: A: PDB Molecule: catalase-peroxidase;PDBTitle: crystal structure of catalase-peroxidase from synechococcus pcc 7942
35 d2ccaa1
not modelled
14.0
33
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG36 d1hwua_
not modelled
13.6
26
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein37 d1e5ma2
not modelled
13.3
12
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related38 d1tqya2
not modelled
13.3
16
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related39 c3ncpD_
not modelled
13.1
16
PDB header: signaling proteinChain: D: PDB Molecule: nitrogen regulatory protein p-ii (glnb-2);PDBTitle: glnk2 from archaeoglobus fulgidus
40 d1vfja_
not modelled
13.0
12
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein41 d1z2la1
not modelled
12.6
14
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases42 d1ul3a_
not modelled
11.6
20
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein43 d1ub2a2
not modelled
10.5
22
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG44 d1r3na1
not modelled
10.3
14
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases45 d1danl3
not modelled
10.3
67
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain46 d1rbli_
not modelled
10.0
17
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit47 c3mk7F_
not modelled
9.7
22
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
48 c2xqoA_
not modelled
9.3
40
PDB header: hydrolaseChain: A: PDB Molecule: cellulosome enzyme, dockerin type i;PDBTitle: ctcel124: a cellulase from clostridium thermocellum
49 c3bzqA_
not modelled
9.1
19
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: nitrogen regulatory protein p-ii;PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
50 d1nh2a1
not modelled
9.1
16
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain51 c2kv5A_
not modelled
9.0
33
PDB header: toxinChain: A: PDB Molecule: putative uncharacterized protein rnai;PDBTitle: solution structure of the par toxin fst in dpc micelles
52 d2gfva2
not modelled
8.9
18
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related53 d2ccaa2
not modelled
8.7
22
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG54 c1yr2A_
not modelled
8.6
21
PDB header: hydrolaseChain: A: PDB Molecule: prolyl oligopeptidase;PDBTitle: structural and mechanistic analysis of two prolyl endopeptidases: role2 of inter-domain dynamics in catalysis and specificity
55 d1u5ta2
not modelled
8.4
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain56 d1oywa1
not modelled
8.1
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RecQ helicase DNA-binding domain-like57 c3bl6A_
not modelled
8.0
10
PDB header: hydrolaseChain: A: PDB Molecule: 5'-methylthioadenosine nucleosidase/s-PDBTitle: crystal structure of staphylococcus aureus 5'-2 methylthioadenosine/s-adenosylhomocysteine nucleosidase in3 complex with formycin a
58 d1itka1
not modelled
7.9
50
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG59 d1ub2a1
not modelled
7.8
33
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG60 d1mwva1
not modelled
7.8
33
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG61 c2oghA_
not modelled
7.7
9
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor eif-1;PDBTitle: solution structure of yeast eif1
62 c3bzjA_
not modelled
7.7
19
PDB header: hydrolaseChain: A: PDB Molecule: uv endonuclease;PDBTitle: uvde k229l
63 d1itka2
not modelled
7.5
23
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG64 d1xmeb2
not modelled
7.4
37
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region65 d1jqna_
not modelled
7.2
16
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase66 c2q3vB_
not modelled
7.0
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein at2g34160;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at2g34160
67 d1v8da_
not modelled
6.8
8
Fold: TTHA0583/YokD-likeSuperfamily: TTHA0583/YokD-likeFamily: Hypothetical protein TT167968 d2juza1
not modelled
6.8
22
Fold: YejL-likeSuperfamily: YejL-likeFamily: YejL-like69 d1cdwa2
not modelled
6.7
23
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain70 d2qtia1
not modelled
6.6
25
Fold: YejL-likeSuperfamily: YejL-likeFamily: YejL-like71 c2imoA_
not modelled
6.6
12
PDB header: hydrolaseChain: A: PDB Molecule: allantoate amidohydrolase;PDBTitle: crystal structure of allantoate amidohydrolase from escherichia coli2 at ph 4.6
72 d1mwva2
not modelled
6.3
22
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG73 d2auna2
not modelled
6.2
26
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like74 c3rrwB_
not modelled
6.2
44
PDB header: plant proteinChain: B: PDB Molecule: thylakoid lumenal 29 kda protein, chloroplastic;PDBTitle: crystal structure of the tl29 protein from arabidopsis thaliana
75 d1uc8a1
not modelled
6.1
32
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Lysine biosynthesis enzyme LysX, N-terminal domain76 d1cdwa1
not modelled
6.1
16
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain77 d2jrxa1
not modelled
6.0
19
Fold: YejL-likeSuperfamily: YejL-likeFamily: YejL-like78 d2cu3a1
not modelled
5.8
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS79 d2v7fa1
not modelled
5.6
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rps19E-like80 c3g23A_
not modelled
5.5
25
PDB header: hydrolaseChain: A: PDB Molecule: ld-carboxypeptidase a;PDBTitle: crystal structure of a ld-carboxypeptidase a (saro_1426) from2 novosphingobium aromaticivorans dsm at 1.89 a resolution
81 c3eyiB_
not modelled
5.5
28
PDB header: dna binding protein/dnaChain: B: PDB Molecule: z-dna-binding protein 1;PDBTitle: the crystal structure of the second z-dna binding domain of2 human dai (zbp1) in complex with z-dna
82 d1jqoa_
not modelled
5.4
20
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase83 c1jqoA_
not modelled
5.4
20
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxylase;PDBTitle: crystal structure of c4-form phosphoenolpyruvate carboxylase from2 maize
84 d2if1a_
not modelled
5.4
26
Fold: eIF1-likeSuperfamily: eIF1-likeFamily: eIF1-like85 c3iumA_
not modelled
5.3
23
PDB header: hydrolaseChain: A: PDB Molecule: prolyl endopeptidase;PDBTitle: appep_wtx opened state
86 d2juwa1
not modelled
5.3
25
Fold: YejL-likeSuperfamily: YejL-likeFamily: YejL-like87 d1j3na2
not modelled
5.3
19
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related88 c2latA_
not modelled
5.2
7
PDB header: membrane proteinChain: A: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: solution structure of a human minimembrane protein ost4
89 c2h3oA_
not modelled
5.2
17
PDB header: membrane proteinChain: A: PDB Molecule: merf;PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
90 c1qysA_
not modelled
5.1
13
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold