| 1 | d1l6sa_
|
|
 |
100.0 |
100 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 2 | d2c1ha1
|
|
 |
100.0 |
47 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 3 | c3obkH_
|
|
 |
100.0 |
43 |
PDB header:lyase
Chain: H: PDB Molecule:delta-aminolevulinic acid dehydratase;
PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
|
| 4 | d1gzga_
|
|
 |
100.0 |
41 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
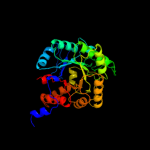
| 5 | d1h7na_
|
|
 |
100.0 |
40 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
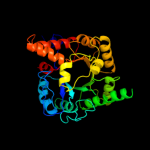
| 6 | d1pv8a_
|
|
 |
100.0 |
41 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
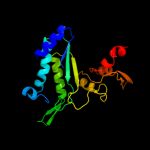
| 7 | d1ps9a1
|
|
 |
97.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
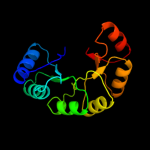
| 8 | d1wbha1
|
|
 |
97.2 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 9 | d1svda1
|
|
 |
96.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 10 | c2h90A_
|
|
 |
96.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
| 11 | c3thaB_
|
|
 |
96.7 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
| 12 | c2qygC_
|
|
 |
96.6 |
21 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
| 13 | d1ykwa1
|
|
 |
96.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 14 | c1ps9A_
|
|
 |
96.6 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 15 | c3eooL_
|
|
 |
96.5 |
21 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 16 | c3nwrA_
|
|
 |
96.4 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
| 17 | c1telA_
|
|
 |
96.4 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase, large subunit;
PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
|
| 18 | c3qfwB_
|
|
 |
96.4 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase large
PDBTitle: crystal structure of rubisco-like protein from rhodopseudomonas2 palustris
|
| 19 | c1rldB_
|
|
 |
96.3 |
17 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose 1,5 bisphosphate carboxylase/oxygenase (large
PDBTitle: solid-state phase transition in the crystal structure of ribulose 1,5-2 biphosphate carboxylase(slash)oxygenase
|
| 20 | c1rcxH_
|
|
 |
96.2 |
17 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
| 21 | d1ej7l1 |
|
not modelled |
96.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 22 | d1mxsa_ |
|
not modelled |
96.2 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 23 | d5ruba1 |
|
not modelled |
96.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 24 | c3fk4A_ |
|
not modelled |
96.0 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:rubisco-like protein;
PDBTitle: crystal structure of rubisco-like protein from bacillus2 cereus atcc 14579
|
| 25 | d1geha1 |
|
not modelled |
96.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 26 | d1bxna1 |
|
not modelled |
95.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 27 | d2d69a1 |
|
not modelled |
95.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 28 | c2zviB_ |
|
not modelled |
95.8 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate
PDBTitle: crystal structure of 2,3-diketo-5-methylthiopentyl-1-2 phosphate enolase from bacillus subtilis
|
| 29 | c2eq5D_ |
|
not modelled |
95.8 |
21 |
PDB header:isomerase
Chain: D: PDB Molecule:228aa long hypothetical hydantoin racemase;
PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
|
| 30 | d1rbla1 |
|
not modelled |
95.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 31 | c1gehE_ |
|
not modelled |
95.7 |
17 |
PDB header:lyase
Chain: E: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase;
PDBTitle: crystal structure of archaeal rubisco (ribulose 1,5-bisphosphate2 carboxylase/oxygenase)
|
| 32 | c2ze3A_ |
|
not modelled |
95.7 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 33 | c2d69B_ |
|
not modelled |
95.7 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: crystal structure of the complex of sulfate ion and octameric2 ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from3 pyrococcus horikoshii ot3 (form-2 crystal)
|
| 34 | c3labA_ |
|
not modelled |
95.6 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate)
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
| 35 | c2oemA_ |
|
not modelled |
95.6 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate enolase;
PDBTitle: crystal structure of a rubisco-like protein from geobacillus2 kaustophilus liganded with mg2+ and 2,3-diketohexane 1-phosphate
|
| 36 | d1muma_ |
|
not modelled |
95.6 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 37 | d1wdda1 |
|
not modelled |
95.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 38 | d1oy0a_ |
|
not modelled |
95.3 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 39 | c3fa4D_ |
|
not modelled |
95.2 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 40 | c3gr7A_ |
|
not modelled |
95.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 41 | d8ruca1 |
|
not modelled |
95.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 42 | d1djqa1 |
|
not modelled |
94.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 43 | c3ez4B_ |
|
not modelled |
94.7 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 44 | c3hf3A_ |
|
not modelled |
94.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 45 | c1zlpA_ |
|
not modelled |
94.5 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 46 | d1gk8a1 |
|
not modelled |
94.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 47 | c2rusB_ |
|
not modelled |
94.4 |
17 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:rubisco (ribulose-1,5-bisphosphate
PDBTitle: crystal structure of the ternary complex of ribulose-1,5-2 bisphosphate carboxylase, mg(ii), and activator co2 at 2.3-3 angstroms resolution
|
| 48 | d1vjia_ |
|
not modelled |
94.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 49 | d1vhca_ |
|
not modelled |
94.2 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 50 | d1bwva1 |
|
not modelled |
94.1 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 51 | d1rd5a_ |
|
not modelled |
94.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 52 | c1djnB_ |
|
not modelled |
93.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trimethylamine dehydrogenase;
PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
|
| 53 | d1m3ua_ |
|
not modelled |
93.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 54 | c9rubB_ |
|
not modelled |
93.2 |
18 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase;
PDBTitle: crystal structure of activated ribulose-1,5-bisphosphate2 carboxylase complexed with its substrate, ribulose-1,5-3 bisphosphate
|
| 55 | c1yadD_ |
|
not modelled |
92.5 |
30 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein teni;
PDBTitle: structure of teni from bacillus subtilis
|
| 56 | c1bwvA_ |
|
not modelled |
92.3 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:protein (ribulose bisphosphate carboxylase);
PDBTitle: activated ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco)2 complexed with the reaction intermediate analogue 2-carboxyarabinitol3 1,5-bisphosphate
|
| 57 | c3kruC_ |
|
not modelled |
92.2 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of the thermostable old yellow enzyme from2 thermoanaerobacter pseudethanolicus e39
|
| 58 | d1wa3a1 |
|
not modelled |
91.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 59 | c2qjhH_ |
|
not modelled |
91.3 |
18 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
| 60 | c3k30B_ |
|
not modelled |
91.1 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
| 61 | c2qiwA_ |
|
not modelled |
91.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 62 | d2czda1 |
|
not modelled |
91.0 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 63 | c2vlbC_ |
|
not modelled |
90.7 |
22 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 64 | d1ujqa_ |
|
not modelled |
90.6 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 65 | d1xcfa_ |
|
not modelled |
90.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 66 | c3gkaB_ |
|
not modelled |
90.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
| 67 | d1s2wa_ |
|
not modelled |
89.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 68 | c1zfjA_ |
|
not modelled |
88.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 69 | c3lerA_ |
|
not modelled |
87.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 70 | c2y7eA_ |
|
not modelled |
87.7 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
|
| 71 | d3bofa2 |
|
not modelled |
87.6 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 72 | c3chvA_ |
|
not modelled |
86.9 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
| 73 | c3cixA_ |
|
not modelled |
86.9 |
17 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
| 74 | c3bolB_ |
|
not modelled |
84.8 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 75 | d1h5ya_ |
|
not modelled |
84.8 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 76 | c3js3C_ |
|
not modelled |
84.6 |
21 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 77 | c3lyeA_ |
|
not modelled |
84.5 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 78 | c1ydoC_ |
|
not modelled |
84.2 |
23 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
| 79 | c2hjpA_ |
|
not modelled |
84.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 80 | c3s5oA_ |
|
not modelled |
83.7 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 81 | d2a6na1 |
|
not modelled |
83.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 82 | c3q58A_ |
|
not modelled |
82.9 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
| 83 | d1gqna_ |
|
not modelled |
81.4 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 84 | c3no5C_ |
|
not modelled |
80.9 |
14 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
|
| 85 | c3ih1A_ |
|
not modelled |
80.5 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 86 | d1vyra_ |
|
not modelled |
79.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 87 | c2ekcA_ |
|
not modelled |
79.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 88 | d1z41a1 |
|
not modelled |
78.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 89 | c3eb2A_ |
|
not modelled |
78.7 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 90 | c2infB_ |
|
not modelled |
78.7 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 bacillus subtilis
|
| 91 | c1rr2A_ |
|
not modelled |
76.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 92 | c3s1vD_ |
|
not modelled |
76.4 |
32 |
PDB header:transferase
Chain: D: PDB Molecule:probable transaldolase;
PDBTitle: transaldolase from thermoplasma acidophilum in complex with d-fructose2 6-phosphate schiff-base intermediate
|
| 93 | c1ydnA_ |
|
not modelled |
76.1 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 94 | d1km4a_ |
|
not modelled |
76.0 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 95 | d1xxxa1 |
|
not modelled |
76.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 96 | c3o63B_ |
|
not modelled |
75.2 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:probable thiamine-phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
|
| 97 | d1gwja_ |
|
not modelled |
75.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 98 | d1o5ka_ |
|
not modelled |
72.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 99 | d1lt7a_ |
|
not modelled |
71.8 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 100 | c3dz1A_ |
|
not modelled |
71.8 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 101 | c3b8iF_ |
|
not modelled |
70.9 |
20 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 102 | c3na8A_ |
|
not modelled |
70.9 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 103 | c3e96B_ |
|
not modelled |
70.1 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 104 | c1x1oC_ |
|
not modelled |
69.5 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
|
| 105 | c3d0cB_ |
|
not modelled |
69.5 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
|
| 106 | c3qfeB_ |
|
not modelled |
68.7 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
| 107 | d1qopa_ |
|
not modelled |
67.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 108 | d1umya_ |
|
not modelled |
67.4 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 109 | d1l6wa_ |
|
not modelled |
66.6 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 110 | d1dvja_ |
|
not modelled |
66.4 |
37 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 111 | c3c6cA_ |
|
not modelled |
66.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
|
| 112 | d1ajza_ |
|
not modelled |
66.2 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 113 | c2ehhE_ |
|
not modelled |
66.0 |
23 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 114 | c2yr1B_ |
|
not modelled |
65.9 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
|
| 115 | c2v9dB_ |
|
not modelled |
65.5 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 116 | c3ct7E_ |
|
not modelled |
64.8 |
17 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
| 117 | c2yw3E_ |
|
not modelled |
64.8 |
24 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
| 118 | d1q45a_ |
|
not modelled |
64.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 119 | d1f74a_ |
|
not modelled |
64.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 120 | d1icpa_ |
|
not modelled |
64.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |