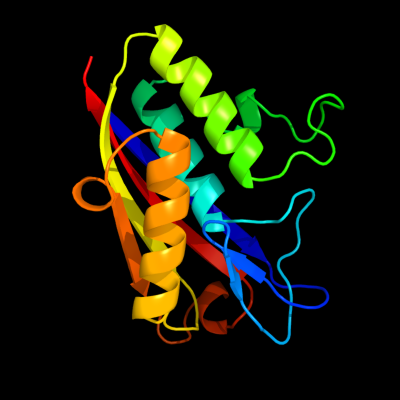
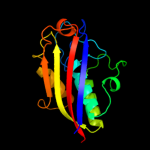
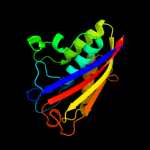
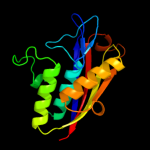
1 c3f0gA_
100.0
63
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: co-crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate2 synthase with cmp
2 d1t0aa_
100.0
67
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like3 d1gx1a_
100.0
97
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like4 c2pmpA_
100.0
48
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from2 the isoprenoid biosynthetic pathway of arabidopsis thaliana
5 d1w55a2
100.0
50
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like6 c1w57A_
100.0
50
PDB header: transferaseChain: A: PDB Molecule: ispd/ispf bifunctional enzyme;PDBTitle: structure of the bifunctional ispdf from campylobacter2 jejuni containing zn
7 d1iv3a_
100.0
41
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like8 d1vh8a_
100.0
77
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like9 c2uzhB_
100.0
40
PDB header: lyaseChain: B: PDB Molecule: 2c-methyl-d-erythritol 2,4-cyclodiphosphatePDBTitle: mycobacterium smegmatis 2c-methyl-d-erythritol-2,4-2 cyclodiphosphate synthase (ispf)
10 c3re3B_
100.0
56
PDB header: lyaseChain: B: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: crystal structure of 2-c-methyl-d-erythritol 2,4-cyclodiphosphate2 synthase from francisella tularensis
11 c3b6nA_
100.0
41
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphatePDBTitle: crystal structure of 2c-methyl-d-erythritol 2,4-2 cyclodiphosphate synthase pv003920 from plasmodium vivax
12 c3fwtA_
64.2
18
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: crystal structure of leishmania major mif2
13 c3b64A_
55.2
17
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: macrophage migration inhibitory factor (mif) from2 /leishmania major
14 d1hfoa_
54.9
14
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related15 d2gdga1
54.5
14
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related16 c1w25B_
53.8
20
PDB header: signaling proteinChain: B: PDB Molecule: stalked-cell differentiation controlling protein;PDBTitle: response regulator pled in complex with c-digmp
17 c2os5C_
52.1
23
PDB header: cytokineChain: C: PDB Molecule: acemif;PDBTitle: macrophage migration inhibitory factor from ancylostoma ceylanicum
18 c2xczA_
50.6
17
PDB header: immune systemChain: A: PDB Molecule: possible atls1-like light-inducible protein;PDBTitle: crystal structure of macrophage migration inhibitory factor2 homologue from prochlorococcus marinus
19 c3bblA_
37.9
22
PDB header: regulatory proteinChain: A: PDB Molecule: regulatory protein of laci family;PDBTitle: crystal structure of a regulatory protein of laci family from2 chloroflexus aggregans
20 d1s5ja2
35.9
10
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: DNA polymerase I21 d1olta_
not modelled
34.0
17
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Oxygen-independent coproporphyrinogen III oxidase HemN22 d1wh9a_
not modelled
29.6
15
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)23 d2cc0a1
not modelled
27.1
13
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase24 d1ru8a_
not modelled
25.5
11
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases25 c3caiA_
not modelled
23.5
16
PDB header: transferaseChain: A: PDB Molecule: possible aminotransferase;PDBTitle: crystal structure of mycobacterium tuberculosis rv3778c2 protein
26 c3hrdE_
not modelled
20.7
26
PDB header: oxidoreductaseChain: E: PDB Molecule: nicotinate dehydrogenase large molybdopterinPDBTitle: crystal structure of nicotinate dehydrogenase
27 c1xaxA_
not modelled
19.9
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0054 protein hi0004;PDBTitle: nmr structure of hi0004, a putative essential gene product2 from haemophilus influenzae
28 d1yh5a1
not modelled
19.8
13
Fold: YggU-likeSuperfamily: YggU-likeFamily: YggU-like29 d1u6ea2
not modelled
19.0
20
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like30 c2ehtA_
not modelled
18.2
29
PDB header: lipid transportChain: A: PDB Molecule: acyl carrier protein;PDBTitle: crystal structure of acyl carrier protein from aquifex aeolicus (form2 2)
31 c3gacD_
not modelled
17.3
16
PDB header: cytokineChain: D: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: structure of mif with hpp
32 d1q8ia2
not modelled
17.2
18
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: DNA polymerase I33 d1h4ra2
not modelled
16.6
23
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM34 c3ejbC_
not modelled
15.5
27
PDB header: oxidoreductase/lipid transportChain: C: PDB Molecule: acyl carrier protein;PDBTitle: crystal structure of p450bioi in complex with tetradecanoic2 acid ligated acyl carrier protein
35 c3izbU_
not modelled
15.4
20
PDB header: ribosomeChain: U: PDB Molecule: 40s ribosomal protein s24;PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
36 c3iz6U_
not modelled
15.4
15
PDB header: ribosomeChain: U: PDB Molecule: 40s ribosomal protein s24 (s24e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
37 d1hnja2
not modelled
15.1
25
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like38 d1tgoa2
not modelled
14.8
18
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: DNA polymerase I39 c2hmaA_
not modelled
14.8
18
PDB header: transferaseChain: A: PDB Molecule: probable trna (5-methylaminomethyl-2-thiouridylate)-PDBTitle: the crystal structure of trna (5-methylaminomethyl-2-thiouridylate)-2 methyltransferase trmu from streptococcus pneumoniae
40 c3t5sA_
not modelled
14.4
8
PDB header: immune systemChain: A: PDB Molecule: macrophage migration inhibitory factor;PDBTitle: structure of macrophage migration inhibitory factor from giardia2 lamblia
41 d2j13a1
not modelled
14.1
29
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase42 d1gyxa_
not modelled
13.2
10
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like43 d1qhta2
not modelled
13.1
18
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: DNA polymerase I44 d1d5aa2
not modelled
13.0
20
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: DNA polymerase I45 d2iboa1
not modelled
13.0
13
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like46 d1n62b2
not modelled
12.8
18
Fold: Molybdenum cofactor-binding domainSuperfamily: Molybdenum cofactor-binding domainFamily: Molybdenum cofactor-binding domain47 c3lvmB_
not modelled
12.6
19
PDB header: transferaseChain: B: PDB Molecule: cysteine desulfurase;PDBTitle: crystal structure of e.coli iscs
48 c1wygA_
not modelled
12.4
24
PDB header: oxidoreductaseChain: A: PDB Molecule: xanthine dehydrogenase/oxidase;PDBTitle: crystal structure of a rat xanthine dehydrogenase triple mutant2 (c535a, c992r and c1324s)
49 c2zkqc_
not modelled
12.2
15
PDB header: ribosomal protein/rnaChain: C: PDB Molecule: rna expansion segment es4;PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
50 d2g1da1
not modelled
12.2
17
Fold: Ribosomal proteins S24e, L23 and L15eSuperfamily: Ribosomal proteins S24e, L23 and L15eFamily: Ribosomal protein S24e51 c2epiA_
not modelled
12.2
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0045 protein mj1052;PDBTitle: crystal structure pf hypothetical protein mj1052 from2 methanocaldococcus jannascii (form 2)
52 d1v97a5
not modelled
12.0
24
Fold: Molybdenum cofactor-binding domainSuperfamily: Molybdenum cofactor-binding domainFamily: Molybdenum cofactor-binding domain53 d1wn7a2
not modelled
11.9
18
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: DNA polymerase I54 c1cmlA_
not modelled
11.5
13
PDB header: transferaseChain: A: PDB Molecule: protein (chalcone synthase);PDBTitle: chalcone synthase from alfalfa complexed with malonyl-coa
55 c2l9fA_
not modelled
11.5
13
PDB header: transferaseChain: A: PDB Molecule: cale8;PDBTitle: nmr solution structure of meacp
56 c2vfjA_
not modelled
11.2
31
PDB header: hydrolaseChain: A: PDB Molecule: tumor necrosis factor;PDBTitle: structure of the a20 ovarian tumour (otu) domain
57 d2c1ia1
not modelled
10.8
19
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase58 c3nctC_
not modelled
10.7
35
PDB header: dna binding protein, chaperoneChain: C: PDB Molecule: protein psib;PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
59 d2fi0a1
not modelled
10.4
26
Fold: SP0561-likeSuperfamily: SP0561-likeFamily: SP0561-like60 c3g8rA_
not modelled
10.4
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: probable spore coat polysaccharide biosynthesis protein e;PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
61 c2fq2A_
not modelled
10.3
17
PDB header: lipid transportChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of minor conformation of holo-acyl2 carrier protein from malaria parasite plasmodium falciparum
62 c3hdoB_
not modelled
10.3
16
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a histidinol-phosphate aminotransferase from2 geobacter metallireducens
63 c3ffhA_
not modelled
10.2
11
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: the crystal structure of histidinol-phosphate aminotransferase from2 listeria innocua clip11262.
64 d1qz9a_
not modelled
10.2
20
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like65 d1ywxa1
not modelled
10.2
11
Fold: Ribosomal proteins S24e, L23 and L15eSuperfamily: Ribosomal proteins S24e, L23 and L15eFamily: Ribosomal protein S24e66 d1eg5a_
not modelled
10.1
23
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like67 c1x3oA_
not modelled
10.0
26
PDB header: lipid transportChain: A: PDB Molecule: acyl carrier protein;PDBTitle: crystal structure of the acyl carrier protein from thermus2 thermophilus hb8
68 c1ffvB_
not modelled
9.8
15
PDB header: hydrolaseChain: B: PDB Molecule: cutl, molybdoprotein of carbon monoxidePDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava
69 c1s5jA_
not modelled
9.7
12
PDB header: transferaseChain: A: PDB Molecule: dna polymerase i;PDBTitle: insight in dna replication: the crystal structure of dna2 polymerase b1 from the archaeon sulfolobus solfataricus
70 d1rm6a2
not modelled
9.6
21
Fold: Molybdenum cofactor-binding domainSuperfamily: Molybdenum cofactor-binding domainFamily: Molybdenum cofactor-binding domain71 c1vlbA_
not modelled
9.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde oxidoreductase;PDBTitle: structure refinement of the aldehyde oxidoreductase from2 desulfovibrio gigas at 1.28 a
72 d1vk8a_
not modelled
9.4
14
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like73 d1ny1a_
not modelled
9.3
28
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase74 c1n62E_
not modelled
9.2
19
PDB header: oxidoreductaseChain: E: PDB Molecule: carbon monoxide dehydrogenase large chain;PDBTitle: crystal structure of the mo,cu-co dehydrogenase (codh), n-2 butylisocyanide-bound state
75 c3dkbA_
not modelled
9.2
31
PDB header: hydrolaseChain: A: PDB Molecule: tumor necrosis factor, alpha-induced protein 3;PDBTitle: crystal structure of a20, 2.5 angstrom
76 c3a2kB_
not modelled
9.2
11
PDB header: ligase/rnaChain: B: PDB Molecule: trna(ile)-lysidine synthase;PDBTitle: crystal structure of tils complexed with trna
77 d1t8ka_
not modelled
9.1
26
Fold: Acyl carrier protein-likeSuperfamily: ACP-likeFamily: Acyl-carrier protein (ACP)78 d1otfa_
not modelled
9.0
29
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like79 c3lkzB_
not modelled
9.0
15
PDB header: viral proteinChain: B: PDB Molecule: non-structural protein 5;PDBTitle: structural and functional analyses of a conserved hydrophobic pocket2 of flavivirus methyltransferase
80 d2p81a1
not modelled
8.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain81 c2op8A_
not modelled
8.8
12
PDB header: isomeraseChain: A: PDB Molecule: probable tautomerase ywhb;PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
82 c1w17A_
not modelled
8.8
28
PDB header: hydrolaseChain: A: PDB Molecule: probable polysaccharide deacetylase pdaa;PDBTitle: structure of bacillus subtilis pdaa, a family 42 carbohydrate esterase.
83 c2hdyA_
not modelled
8.7
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: selenocysteine lyase;PDBTitle: structure of human selenocysteine lyase
84 c3ry0A_
not modelled
8.6
26
PDB header: isomeraseChain: A: PDB Molecule: putative tautomerase;PDBTitle: crystal structure of tomn, a 4-oxalocrotonate tautomerase homologue in2 tomaymycin biosynthetic pathway
85 d1elua_
not modelled
8.6
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like86 d1s6ya2
not modelled
8.5
24
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase87 d1yqha1
not modelled
8.4
19
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like88 d1xn9a_
not modelled
8.4
17
Fold: Ribosomal proteins S24e, L23 and L15eSuperfamily: Ribosomal proteins S24e, L23 and L15eFamily: Ribosomal protein S24e89 c3m20A_
not modelled
8.3
20
PDB header: isomeraseChain: A: PDB Molecule: 4-oxalocrotonate tautomerase, putative;PDBTitle: crystal structure of dmpi from archaeoglobus fulgidus determined to2 2.37 angstroms resolution
90 d1ub7a2
not modelled
8.3
21
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like91 c2oq2B_
not modelled
8.2
11
PDB header: oxidoreductaseChain: B: PDB Molecule: phosphoadenosine phosphosulfate reductase;PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
92 d3clsc1
not modelled
8.0
17
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits93 c3lmoA_
not modelled
8.0
17
PDB header: transferaseChain: A: PDB Molecule: specialized acyl carrier protein;PDBTitle: crystal structure of specialized acyl carrier protein2 (rpa2022) from rhodopseudomonas palustris, northeast3 structural genomics consortium target rpr324
94 c3pfyA_
not modelled
7.9
31
PDB header: hydrolaseChain: A: PDB Molecule: otu domain-containing protein 5;PDBTitle: the catalytic domain of human otud5
95 d1mzja2
not modelled
7.5
18
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like96 d1r57a_
not modelled
7.5
10
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT97 c1sb3D_
not modelled
7.5
21
PDB header: oxidoreductaseChain: D: PDB Molecule: 4-hydroxybenzoyl-coa reductase alpha subunit;PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
98 c2bg5C_
not modelled
7.4
19
PDB header: transferaseChain: C: PDB Molecule: phosphoenolpyruvate-protein kinase;PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme2 i-domain from the thermoanaerobacter tengcongensis pep:3 sugar phosphotransferase system (pts)
99 d1lc5a_
not modelled
7.3
10
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like