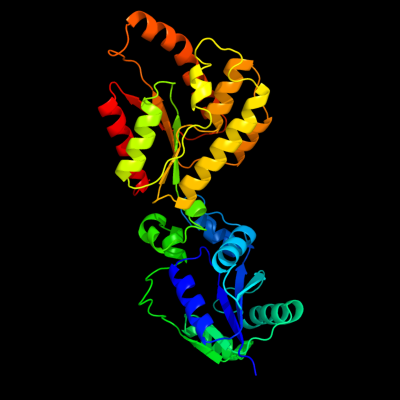
| 1 | c1lw7A_
|
|
 |
100.0 |
53 |
PDB header:transferase
Chain: A: PDB Molecule:transcriptional regulator nadr;
PDBTitle: nadr protein from haemophilus influenzae
|
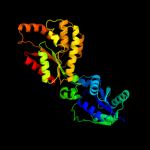
| 2 | d1lw7a2
|
|
 |
100.0 |
56 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
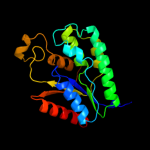
| 3 | d1lw7a1
|
|
 |
100.0 |
51 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 4 | c2qjoB_
|
|
 |
99.9 |
17 |
PDB header:transferase, hydrolase
Chain: B: PDB Molecule:bifunctional nmn adenylyltransferase/nudix hydrolase;
PDBTitle: crystal structure of a bifunctional nmn adenylyltransferase/adp ribose2 pyrophosphatase (nadm) complexed with adprp and nad from3 synechocystis sp.
|
| 5 | d1f9aa_
|
|
 |
99.9 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 6 | c3f3mA_
|
|
 |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: six crystal structures of two phosphopantetheine2 adenylyltransferases reveal an alternative ligand binding3 mode and an associated structural change
|
| 7 | d1ej2a_
|
|
 |
99.9 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 8 | d1qjca_
|
|
 |
99.9 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 9 | d1tfua_
|
|
 |
99.9 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 10 | c3ikzA_
|
|
 |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: crystal structure of phosphopantetheine adenylyltransferase from2 burkholderia pseudomallei
|
| 11 | d1o6ba_
|
|
 |
99.9 |
18 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 12 | d1vlha_
|
|
 |
99.9 |
22 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 13 | c2r5wA_
|
|
 |
99.9 |
17 |
PDB header:hydrolase, transferase
Chain: A: PDB Molecule:nicotinamide-nucleotide adenylyltransferase;
PDBTitle: crystal structure of a bifunctional nmn2 adenylyltransferase/adp ribose pyrophosphatase from3 francisella tularensis
|
| 14 | c3op1A_
|
|
 |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:macrolide-efflux protein;
PDBTitle: crystal structure of macrolide-efflux protein sp_1110 from2 streptococcus pneumoniae
|
| 15 | c3nd5D_
|
|
 |
99.9 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: crystal structure of phosphopantetheine adenylyltransferase (ppat)2 from enterococcus faecalis
|
| 16 | c3nv7A_
|
|
 |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: crystal structure of h.pylori phosphopantetheine adenylyltransferase2 mutant i4v/n76y
|
| 17 | c2x0kB_
|
|
 |
99.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin biosynthesis protein ribf;
PDBTitle: crystal structure of modular fad synthetase from2 corynebacterium ammoniagenes
|
| 18 | c3e27B_
|
|
 |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:nicotinate (nicotinamide) nucleotide
PDBTitle: nicotinic acid mononucleotide (namn) adenylyltransferase2 from bacillus anthracis: product complex
|
| 19 | d1od6a_
|
|
 |
99.9 |
18 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 20 | d1kama_
|
|
 |
99.9 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 21 | c2h29A_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide
PDBTitle: crystal structure of nicotinic acid mononucleotide2 adenylyltransferase from staphylococcus aureus: product3 bound form 1
|
| 22 | d1k4ma_ |
|
not modelled |
99.8 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 23 | c1yunB_ |
|
not modelled |
99.8 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:probable nicotinate-nucleotide
PDBTitle: crystal structure of nicotinic acid mononucleotide2 adenylyltransferase from pseudomonas aeruginosa
|
| 24 | d1nuua_ |
|
not modelled |
99.8 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 25 | d1kr2a_ |
|
not modelled |
99.8 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 26 | c1t6zB_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin kinase/fmn adenylyltransferase;
PDBTitle: crystal structure of riboflavin bound tm379
|
| 27 | c3h05A_ |
|
not modelled |
99.8 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vpa0413;
PDBTitle: the crystal structure of a putative nicotinate-nucleotide2 adenylyltransferase from vibrio parahaemolyticus
|
| 28 | d1mrza2 |
|
not modelled |
99.8 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 29 | d1tmka_ |
|
not modelled |
99.7 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 30 | d1coza_ |
|
not modelled |
99.7 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Cytidylyltransferase |
| 31 | c3ld9D_ |
|
not modelled |
99.6 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:thymidylate kinase;
PDBTitle: crystal structure of thymidylate kinase from ehrlichia chaffeensis at2 2.15a resolution
|
| 32 | c2w0sB_ |
|
not modelled |
99.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:thymidylate kinase;
PDBTitle: crystal structure of vaccinia virus thymidylate kinase2 bound to brivudin-5'-monophosphate
|
| 33 | c3cr8C_ |
|
not modelled |
99.6 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:sulfate adenylyltranferase, adenylylsulfate
PDBTitle: hexameric aps kinase from thiobacillus denitrificans
|
| 34 | d1nn5a_ |
|
not modelled |
99.5 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 35 | c1m8pB_ |
|
not modelled |
99.5 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:sulfate adenylyltransferase;
PDBTitle: crystal structure of p. chrysogenum atp sulfurylase in the t-state
|
| 36 | c2gksB_ |
|
not modelled |
99.5 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional sat/aps kinase;
PDBTitle: crystal structure of the bi-functional atp sulfurylase-aps kinase from2 aquifex aeolicus, a chemolithotrophic thermophile
|
| 37 | c2wwiC_ |
|
not modelled |
99.5 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:thymidilate kinase, putative;
PDBTitle: plasmodium falciparum thymidylate kinase in complex with2 aztmp and adp
|
| 38 | c3glvB_ |
|
not modelled |
99.5 |
20 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:lipopolysaccharide core biosynthesis protein;
PDBTitle: crystal structure of the lipopolysaccharide core biosynthesis protein2 from thermoplasma volcanium gss1
|
| 39 | c2z0hA_ |
|
not modelled |
99.5 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:thymidylate kinase;
PDBTitle: crystal structure of thymidylate kinase in complex with dtdp2 and adp from thermotoga maritima
|
| 40 | d4tmka_ |
|
not modelled |
99.5 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 41 | c3lv8A_ |
|
not modelled |
99.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:thymidylate kinase;
PDBTitle: 1.8 angstrom resolution crystal structure of a thymidylate kinase2 (tmk) from vibrio cholerae o1 biovar eltor str. n16961 in complex3 with tmp, thymidine-5'-diphosphate and adp
|
| 42 | c2ccjA_ |
|
not modelled |
99.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:thymidylate kinase;
PDBTitle: crystal structure of s. aureus thymidylate kinase complexed2 with thymidine monophosphate
|
| 43 | c2plrB_ |
|
not modelled |
99.4 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:probable thymidylate kinase;
PDBTitle: crystal structure of dtmp kinase (st1543) from sulfolobus tokodaii2 strain7
|
| 44 | d1p5zb_ |
|
not modelled |
99.3 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 45 | c2pbrB_ |
|
not modelled |
99.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:thymidylate kinase;
PDBTitle: crystal structure of thymidylate kinase (aq_969) from aquifex aeolicus2 vf5
|
| 46 | c2jatA_ |
|
not modelled |
99.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:deoxyguanosine kinase;
PDBTitle: structure of deoxyadenosine kinase from m.mycoides with2 products dcmp and a flexible dcdp bound
|
| 47 | d1gsia_ |
|
not modelled |
99.3 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 48 | c2qroB_ |
|
not modelled |
99.3 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:deoxycytidine kinase;
PDBTitle: human deoxycytidine kinase damp, udp, mg ion product complex
|
| 49 | c2b7lD_ |
|
not modelled |
99.2 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:glycerol-3-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ctp:glycerol-3-phosphate2 cytidylyltransferase from staphylococcus aureus
|
| 50 | c3elbA_ |
|
not modelled |
99.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:ethanolamine-phosphate cytidylyltransferase;
PDBTitle: human ctp: phosphoethanolamine cytidylyltransferase in complex with2 cmp
|
| 51 | c3guzB_ |
|
not modelled |
99.2 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: structural and substrate-binding studies of pantothenate2 synthenate (ps)provide insights into homotropic inhibition3 by pantoate in ps's
|
| 52 | d2ocpa1 |
|
not modelled |
99.1 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 53 | c1g8gB_ |
|
not modelled |
99.1 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:sulfate adenylyltransferase;
PDBTitle: atp sulfurylase from s. cerevisiae: the binary product complex with2 aps
|
| 54 | d1zaka1 |
|
not modelled |
99.1 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 55 | c2ql6H_ |
|
not modelled |
99.0 |
14 |
PDB header:signaling protein,transferase
Chain: H: PDB Molecule:nicotinamide riboside kinase 1;
PDBTitle: human nicotinamide riboside kinase (nrk1)
|
| 56 | c3gmiA_ |
|
not modelled |
99.0 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0348 protein mj0951;
PDBTitle: crystal structure of a protein of unknown function from2 methanocaldococcus jannaschii
|
| 57 | c1zuiA_ |
|
not modelled |
99.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:shikimate kinase;
PDBTitle: structural basis for shikimate-binding specificity of helicobacter2 pylori shikimate kinase
|
| 58 | d2cdna1 |
|
not modelled |
99.0 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 59 | d1knqa_ |
|
not modelled |
98.9 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Gluconate kinase |
| 60 | d1khta_ |
|
not modelled |
98.9 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 61 | c2pt5D_ |
|
not modelled |
98.9 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:shikimate kinase;
PDBTitle: crystal structure of shikimate kinase (aq_2177) from aquifex aeolicus2 vf5
|
| 62 | c2bwjC_ |
|
not modelled |
98.9 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:adenylate kinase 5;
PDBTitle: structure of adenylate kinase 5
|
| 63 | d3adka_ |
|
not modelled |
98.9 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 64 | d1ak2a1 |
|
not modelled |
98.9 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 65 | d1e4va1 |
|
not modelled |
98.9 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 66 | d1e2ka_ |
|
not modelled |
98.9 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 67 | d1akya1 |
|
not modelled |
98.9 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 68 | c1zakB_ |
|
not modelled |
98.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:adenylate kinase;
PDBTitle: adenylate kinase from maize in complex with the inhibitor2 p1,p5-bis(adenosine-5'-)pentaphosphate (ap5a)
|
| 69 | d2ak3a1 |
|
not modelled |
98.9 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 70 | c2if2C_ |
|
not modelled |
98.9 |
8 |
PDB header:transferase
Chain: C: PDB Molecule:dephospho-coa kinase;
PDBTitle: crystal structure of the putative dephospho-coa kinase from aquifex2 aeolicus, northeast structural genomics target qr72.
|
| 71 | d1nksa_ |
|
not modelled |
98.9 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 72 | c3do8B_ |
|
not modelled |
98.9 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: the crystal structure of the protein with unknown function2 from archaeoglobus fulgidus
|
| 73 | d1p3ja1 |
|
not modelled |
98.9 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 74 | d1ki9a_ |
|
not modelled |
98.8 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 75 | d2vp4a1 |
|
not modelled |
98.8 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 76 | d1teva_ |
|
not modelled |
98.8 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 77 | c3tlxA_ |
|
not modelled |
98.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase 2;
PDBTitle: crystal structure of pf10_0086, adenylate kinase from plasmodium2 falciparum
|
| 78 | d1zina1 |
|
not modelled |
98.8 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 79 | c2eu8B_ |
|
not modelled |
98.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of a thermostable mutant of bacillus2 subtilis adenylate kinase (q199r)
|
| 80 | d1e6ca_ |
|
not modelled |
98.8 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 81 | d1p6xa_ |
|
not modelled |
98.8 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 82 | c3nwjB_ |
|
not modelled |
98.8 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:atsk2;
PDBTitle: crystal structure of shikimate kinase from arabidopsis thaliana2 (atsk2)
|
| 83 | d1s3ga1 |
|
not modelled |
98.8 |
9 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 84 | c2rh5B_ |
|
not modelled |
98.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:adenylate kinase;
PDBTitle: structure of apo adenylate kinase from aquifex aeolicus
|
| 85 | d1viaa_ |
|
not modelled |
98.7 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 86 | d1ukza_ |
|
not modelled |
98.7 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 87 | d1rkba_ |
|
not modelled |
98.7 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 88 | d2bdta1 |
|
not modelled |
98.7 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Atu3015-like |
| 89 | c3hl4B_ |
|
not modelled |
98.7 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:choline-phosphate cytidylyltransferase a;
PDBTitle: crystal structure of a mammalian ctp:phosphocholine2 cytidylyltransferase with cdp-choline
|
| 90 | d1qf9a_ |
|
not modelled |
98.7 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 91 | c2ak3B_ |
|
not modelled |
98.7 |
15 |
PDB header:transferase (phosphotransferase)
Chain: B: PDB Molecule:adenylate kinase isoenzyme-3;
PDBTitle: the three-dimensional structure of the complex between2 mitochondrial matrix adenylate kinase and its substrate3 amp at 1.85 angstroms resolution
|
| 92 | c3akcA_ |
|
not modelled |
98.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:cytidylate kinase;
PDBTitle: crystal structure of cmp kinase in complex with cdp and adp from2 thermus thermophilus hb8
|
| 93 | c3cm0A_ |
|
not modelled |
98.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of adenylate kinase from thermus2 thermophilus hb8
|
| 94 | d1kaga_ |
|
not modelled |
98.7 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 95 | c1ankA_ |
|
not modelled |
98.7 |
10 |
PDB header:transferase(phosphotransferase)
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: the closed conformation of a highly flexible protein: the2 structure of e. coli adenylate kinase with bound amp and3 amppnp
|
| 96 | c2ar7A_ |
|
not modelled |
98.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase 4;
PDBTitle: crystal structure of human adenylate kinase 4, ak4
|
| 97 | c3t61A_ |
|
not modelled |
98.6 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:gluconokinase;
PDBTitle: crystal structure of a gluconokinase from sinorhizobium meliloti 1021
|
| 98 | c2vliB_ |
|
not modelled |
98.6 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:antibiotic resistance protein;
PDBTitle: structure of deinococcus radiodurans tunicamycin resistance2 protein
|
| 99 | d1y63a_ |
|
not modelled |
98.6 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 100 | c2yvuA_ |
|
not modelled |
98.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:probable adenylyl-sulfate kinase;
PDBTitle: crystal structure of ape1195
|
| 101 | c3akyA_ |
|
not modelled |
98.6 |
15 |
PDB header:adenylate kinase
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: stability, activity and structure of adenylate kinase2 mutants
|
| 102 | c1s3gA_ |
|
not modelled |
98.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of adenylate kinase from bacillus2 globisporus
|
| 103 | d2iyva1 |
|
not modelled |
98.5 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 104 | c3h0kA_ |
|
not modelled |
98.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0200 protein sso1041;
PDBTitle: crystal structure of an adenylated kinase related protein from2 sulfolobus solfataricus to 3.25a
|
| 105 | c1qhiA_ |
|
not modelled |
98.5 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:protein (thymidine kinase);
PDBTitle: herpes simplex virus type-i thymidine kinase complexed with2 a novel non-substrate inhibitor, 9-(4-hydroxybutyl)-n2-3 phenylguanine
|
| 106 | d1ckea_ |
|
not modelled |
98.5 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 107 | c2vp4D_ |
|
not modelled |
98.5 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:deoxynucleoside kinase;
PDBTitle: structural studies of nucleoside analog and feedback2 inhibitor binding to drosophila melanogaster3 multisubstrate deoxyribonucleoside kinase
|
| 108 | d1q3ta_ |
|
not modelled |
98.5 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 109 | c2rhmD_ |
|
not modelled |
98.5 |
12 |
PDB header:unknown function
Chain: D: PDB Molecule:putative kinase;
PDBTitle: crystal structure of a putative kinase (caur_3907) from chloroflexus2 aurantiacus j-10-fl at 1.70 a resolution
|
| 110 | c3trfB_ |
|
not modelled |
98.5 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:shikimate kinase;
PDBTitle: structure of a shikimate kinase (arok) from coxiella burnetii
|
| 111 | c2ak2A_ |
|
not modelled |
98.5 |
18 |
PDB header:phosphotransferase
Chain: A: PDB Molecule:adenylate kinase isoenzyme-2;
PDBTitle: adenylate kinase isoenzyme-2
|
| 112 | d1vhta_ |
|
not modelled |
98.4 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 113 | d1gkya_ |
|
not modelled |
98.4 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 114 | d1m7gb_ |
|
not modelled |
98.4 |
9 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Adenosine-5'phosphosulfate kinase (APS kinase) |
| 115 | d1x6va3 |
|
not modelled |
98.4 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Adenosine-5'phosphosulfate kinase (APS kinase) |
| 116 | d1uj2a_ |
|
not modelled |
98.4 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Phosphoribulokinase/pantothenate kinase |
| 117 | c3tr0A_ |
|
not modelled |
98.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:guanylate kinase;
PDBTitle: structure of guanylate kinase (gmk) from coxiella burnetii
|
| 118 | d1m8pa3 |
|
not modelled |
98.3 |
10 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ATP sulfurylase C-terminal domain |
| 119 | c1ueiB_ |
|
not modelled |
98.3 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:uridine-cytidine kinase 2;
PDBTitle: crystal structure of human uridine-cytidine kinase 22 complexed with a feedback-inhibitor, utp
|
| 120 | d1m7ga_ |
|
not modelled |
98.3 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Adenosine-5'phosphosulfate kinase (APS kinase) |