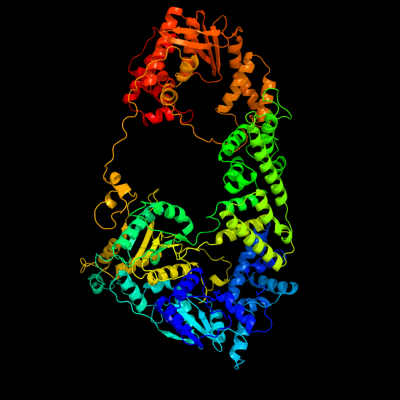
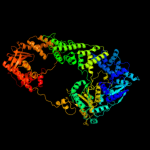
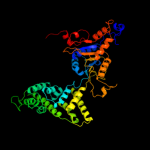
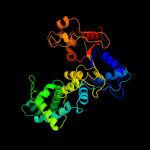
1 c1w36F_
100.0
100
PDB header: recombinationChain: F: PDB Molecule: exodeoxyribonuclease v gamma chain;PDBTitle: recbcd:dna complex
2 d1w36c2
100.0
100
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain3 d1w36c1
100.0
100
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain4 d1w36c3
100.0
100
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Exodeoxyribonuclease V beta chain (RecC), C-terminal domain5 d1uaaa2
100.0
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain6 c2is6B_
100.0
15
PDB header: hydrolase/dnaChain: B: PDB Molecule: dna helicase ii;PDBTitle: crystal structure of uvrd-dna-adpmgf3 ternary complex
7 c1uaaB_
100.0
14
PDB header: hydrolase/dnaChain: B: PDB Molecule: protein (atp-dependent dna helicase rep.);PDBTitle: e. coli rep helicase/dna complex
8 d1pjra2
100.0
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain9 c1w36E_
100.0
13
PDB header: recombinationChain: E: PDB Molecule: exodeoxyribonuclease v beta chain;PDBTitle: recbcd:dna complex
10 c3lfuA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: dna helicase ii;PDBTitle: crystal structure of e. coli uvrd
11 d1w36b2
99.9
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain12 c1pjrA_
99.9
13
PDB header: helicaseChain: A: PDB Molecule: pcra;PDBTitle: structure of dna helicase
13 c2pjrF_
99.8
13
PDB header: hydrolase/dnaChain: F: PDB Molecule: protein (helicase pcra);PDBTitle: helicase product complex
14 c1qhhB_
99.6
10
PDB header: hydrolaseChain: B: PDB Molecule: protein (pcra (subunit));PDBTitle: structure of dna helicase with adpnp
15 c2pjrB_
99.2
23
PDB header: hydrolase/dnaChain: B: PDB Molecule: protein (helicase pcra);PDBTitle: helicase product complex
16 c1qhhD_
99.1
23
PDB header: hydrolaseChain: D: PDB Molecule: protein (pcra (subunit));PDBTitle: structure of dna helicase with adpnp
17 c3dmnA_
98.7
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative dna helicase;PDBTitle: the crystal structure of the c-terminal domain of a possilbe dna2 helicase from lactobacillus plantarun wcfs1
18 d1w36d2
98.0
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain19 c1w36G_
95.3
18
PDB header: recombinationChain: G: PDB Molecule: exodeoxyribonuclease v alpha chain;PDBTitle: recbcd:dna complex
20 c3e1sA_
94.9
16
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease v, subunit recd;PDBTitle: structure of an n-terminal truncation of deinococcus radiodurans recd2
21 c2gk7A_
not modelled
91.5
22
PDB header: hydrolaseChain: A: PDB Molecule: regulator of nonsense transcripts 1;PDBTitle: structural and functional insights into the human upf1 helicase core
22 c2xzlA_
not modelled
82.6
17
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent helicase nam7;PDBTitle: upf1-rna complex
23 c2wjyA_
not modelled
74.0
18
PDB header: hydrolaseChain: A: PDB Molecule: regulator of nonsense transcripts 1;PDBTitle: crystal structure of the complex between human nonsense2 mediated decay factors upf1 and upf2 orthorhombic form
24 d1zrra1
not modelled
61.8
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Acireductone dioxygenase25 d1vr3a1
not modelled
61.2
21
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Acireductone dioxygenase26 c2l2qA_
not modelled
56.1
6
PDB header: transferaseChain: A: PDB Molecule: pts system, cellobiose-specific iib component (cela);PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
27 d1vzva_
not modelled
51.0
17
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin28 c3nbmA_
not modelled
39.5
11
PDB header: transferaseChain: A: PDB Molecule: pts system, lactose-specific iibc components;PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
29 c3h4rA_
not modelled
30.5
10
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease 8;PDBTitle: crystal structure of e. coli rece exonuclease
30 d1at3a_
not modelled
30.5
19
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin31 d1etea_
not modelled
30.0
21
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Short-chain cytokines32 c3nctC_
not modelled
19.8
22
PDB header: dna binding protein, chaperoneChain: C: PDB Molecule: protein psib;PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
33 d1wnaa1
not modelled
19.5
28
Fold: TTHA1528-likeSuperfamily: TTHA1528-likeFamily: TTHA1528-like34 c1qhhA_
not modelled
18.6
13
PDB header: hydrolaseChain: A: PDB Molecule: protein (pcra (subunit));PDBTitle: structure of dna helicase with adpnp
35 d1t1ea2
not modelled
17.2
17
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors36 d2o8ra3
not modelled
15.4
11
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain37 d1l5wa_
not modelled
14.8
18
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Oligosaccharide phosphorylase38 d1p2fa1
not modelled
14.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like39 c3edyA_
not modelled
14.3
14
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure of the precursor form of human tripeptidyl-peptidase2 1
40 c3l0aA_
not modelled
14.3
4
PDB header: hydrolaseChain: A: PDB Molecule: putative exonuclease;PDBTitle: crystal structure of putative exonuclease (rer070207002219) from2 eubacterium rectale at 2.19 a resolution
41 d2bcqa3
not modelled
14.2
15
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like42 c1eg4A_
not modelled
13.6
12
PDB header: structural proteinChain: A: PDB Molecule: dystrophin;PDBTitle: structure of a dystrophin ww domain fragment in complex2 with a beta-dystroglycan peptide
43 c1dipA_
not modelled
12.4
29
PDB header: acetylationChain: A: PDB Molecule: delta-sleep-inducing peptide immunoreactivePDBTitle: the solution structure of porcine delta-sleep-inducing2 peptide immunoreactive peptide, nmr, 10 structures
44 d2cfua2
not modelled
11.5
17
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Alkylsulfatase-like45 d2vo9a1
not modelled
11.4
16
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanY-like46 c1oywA_
not modelled
11.2
7
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase;PDBTitle: structure of the recq catalytic core
47 c2zxxA_
not modelled
11.2
29
PDB header: cell cycle/replicationChain: A: PDB Molecule: geminin;PDBTitle: crystal structure of cdt1/geminin complex
48 d2h5na1
not modelled
11.0
21
Fold: TerB-likeSuperfamily: TerB-likeFamily: PG1108-like49 d1x52a1
not modelled
10.9
33
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like50 d2guka1
not modelled
10.5
13
Fold: PG1857-likeSuperfamily: PG1857-likeFamily: PG1857-like51 d1pjqa3
not modelled
10.5
12
Fold: Siroheme synthase middle domains-likeSuperfamily: Siroheme synthase middle domains-likeFamily: Siroheme synthase middle domains-like52 d1lb2b_
not modelled
10.4
14
Fold: SAM domain-likeSuperfamily: C-terminal domain of RNA polymerase alpha subunitFamily: C-terminal domain of RNA polymerase alpha subunit53 d1oywa2
not modelled
10.2
7
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain54 c2vo9C_
not modelled
10.1
18
PDB header: hydrolaseChain: C: PDB Molecule: l-alanyl-d-glutamate peptidase;PDBTitle: crystal structure of the enzymatically active domain of the2 listeria monocytogenes bacteriophage 500 endolysin ply500
55 c3qy9C_
not modelled
10.0
13
PDB header: oxidoreductaseChain: C: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: the crystal structure of dihydrodipicolinate reductase from2 staphylococcus aureus
56 d1pjra1
not modelled
10.0
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain57 d1iiba_
not modelled
9.9
16
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Lactose/Cellobiose specific IIB subunit58 c3dmqA_
not modelled
9.7
20
PDB header: hydrolaseChain: A: PDB Molecule: rna polymerase-associated protein rapa;PDBTitle: crystal structure of rapa, a swi2/snf2 protein that2 recycles rna polymerase during transcription
59 c2v1xB_
not modelled
9.6
14
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent dna helicase q1;PDBTitle: crystal structure of human recq-like dna helicase
60 c3agjD_
not modelled
9.4
20
PDB header: translation/hydrolaseChain: D: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
61 c3agjB_
not modelled
9.4
20
PDB header: translation/hydrolaseChain: B: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
62 d1tdja3
not modelled
9.4
22
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Allosteric threonine deaminase C-terminal domain63 c1vjqB_
not modelled
9.3
20
PDB header: structural genomics, de novo proteinChain: B: PDB Molecule: designed protein;PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
64 c2d1uA_
not modelled
9.2
26
PDB header: metal transportChain: A: PDB Molecule: iron(iii) dicitrate transport protein feca;PDBTitle: solution strcuture of the periplasmic signaling domain of2 feca from escherichia coli
65 c1w5cT_
not modelled
9.0
23
PDB header: photosynthesisChain: T: PDB Molecule: cytochrome c-550;PDBTitle: photosystem ii from thermosynechococcus elongatus
66 d1dt9a3
not modelled
9.0
13
Fold: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1Superfamily: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1Family: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF167 d2gj4a1
not modelled
8.9
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Oligosaccharide phosphorylase68 d2vgna3
not modelled
8.9
27
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like69 c3i7tA_
not modelled
8.7
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of rv2704, a member of highly conserved2 yjgf/yer057c/uk114 family, from mycobacterium tuberculosis
70 d1qzma_
not modelled
8.6
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain71 c3ijpA_
not modelled
8.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase from2 bartonella henselae at 2.0a resolution
72 c2cfuA_
not modelled
8.6
20
PDB header: hydrolaseChain: A: PDB Molecule: sdsa1;PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
73 c2wvrB_
not modelled
8.4
29
PDB header: replicationChain: B: PDB Molecule: geminin;PDBTitle: human cdt1:geminin complex
74 d1v6ta_
not modelled
8.4
18
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like75 c3ddsB_
not modelled
8.2
15
PDB header: transferaseChain: B: PDB Molecule: glycogen phosphorylase, liver form;PDBTitle: crystal structure of glycogen phosphorylase complexed with an2 anthranilimide based inhibitor gsk261
76 d1kgsa1
not modelled
8.0
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like77 c2pmuD_
not modelled
8.0
14
PDB header: transcription regulationChain: D: PDB Molecule: response regulator phop;PDBTitle: crystal structure of the dna-binding domain of phop
78 c1t1eA_
not modelled
7.7
16
PDB header: hydrolaseChain: A: PDB Molecule: kumamolisin;PDBTitle: high resolution crystal structure of the intact pro-2 kumamolisin, a sedolisin type proteinase (previously3 called kumamolysin or kscp)
79 c3ee6A_
not modelled
7.7
16
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure analysis of tripeptidyl peptidase -i
80 d2fcla1
not modelled
7.7
19
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: TM1012-like81 c3e20C_
not modelled
7.7
12
PDB header: translationChain: C: PDB Molecule: eukaryotic peptide chain release factor subunit 1;PDBTitle: crystal structure of s.pombe erf1/erf3 complex
82 d2pbka1
not modelled
7.6
26
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin83 c2o8rA_
not modelled
7.6
11
PDB header: transferaseChain: A: PDB Molecule: polyphosphate kinase;PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
84 c3lmeE_
not modelled
7.5
24
PDB header: translationChain: E: PDB Molecule: possible translation initiation inhibitor;PDBTitle: structure of probable translation initiation inhibitor from2 (rpa2473) from rhodopseudomonas palustris
85 d1vm6a3
not modelled
7.4
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain86 c1p65A_
not modelled
7.3
24
PDB header: viral proteinChain: A: PDB Molecule: nucleocapsid protein;PDBTitle: crystal structure of the nucleocapsid protein of porcine reproductive2 and respiratory syndrome virus (prrsv)
87 d1p65a_
not modelled
7.3
24
Fold: Nucleocapsid protein dimerization domainSuperfamily: Nucleocapsid protein dimerization domainFamily: Arterivirus nucleocapsid protein88 d2aala1
not modelled
7.2
20
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MSAD-like89 d1nrga_
not modelled
6.9
12
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like90 c1nrgA_
not modelled
6.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: pyridoxine 5'-phosphate oxidase;PDBTitle: structure and properties of recombinant human pyridoxine-5'-phosphate2 oxidase
91 d1mz4a_
not modelled
6.8
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c92 d1cooa_
not modelled
6.7
13
Fold: SAM domain-likeSuperfamily: C-terminal domain of RNA polymerase alpha subunitFamily: C-terminal domain of RNA polymerase alpha subunit93 c1um9D_
not modelled
6.7
22
PDB header: oxidoreductaseChain: D: PDB Molecule: 2-oxo acid dehydrogenase beta subunit;PDBTitle: branched-chain 2-oxo acid dehydrogenase (e1) from thermus2 thermophilus hb8 in apo-form
94 c2vgmA_
not modelled
6.7
27
PDB header: cell cycleChain: A: PDB Molecule: dom34;PDBTitle: structure of yeast dom34 : a protein related to translation2 termination factor erf1 and involved in no-go decay.
95 d2ns0a1
not modelled
6.7
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like96 c2ph0A_
not modelled
6.7
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q6d2t7_erwct protein from erwinia2 carotovora. nesg target ewr41.
97 c1xdoB_
not modelled
6.6
11
PDB header: transferaseChain: B: PDB Molecule: polyphosphate kinase;PDBTitle: crystal structure of escherichia coli polyphosphate kinase
98 c2qrvC_
not modelled
6.6
14
PDB header: transferase/transferase regulatorChain: C: PDB Molecule: dna (cytosine-5)-methyltransferase 3-like;PDBTitle: structure of dnmt3a-dnmt3l c-terminal domain complex
99 d1uaaa1
not modelled
6.6
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain