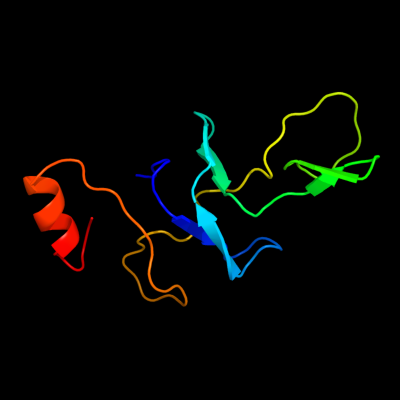
1 d2gycs1
100.0
100
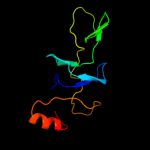
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e2 d2zjrr1
100.0
43
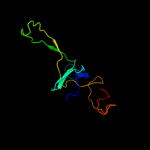
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e3 c3bboW_
100.0
34
PDB header: ribosomeChain: W: PDB Molecule: ribosomal protein l24;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
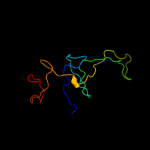
4 d2j01y1
100.0
43
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e5 c2ftcN_
100.0
30
PDB header: ribosomeChain: N: PDB Molecule: mitochondrial ribosomal protein l24;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
6 c3iz5Y_
99.9
28
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
7 d1vqot1
99.9
27
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e8 c4a1cS_
99.9
32
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
9 c1s1iU_
99.8
29
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l26-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
10 c2zkrt_
99.2
33
PDB header: ribosomal protein/rnaChain: T: PDB Molecule: rna expansion segment es39 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
11 c2kvqG_
96.2
19
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
12 c2jvvA_
96.2
19
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
13 d1nppa2
96.0
25
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain14 c3p8bB_
95.7
28
PDB header: transferase/transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
15 d1nz9a_
95.5
31
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain16 d2joya1
95.3
24
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L14e17 d2do3a1
95.0
28
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like18 c2ckkA_
94.0
16
PDB header: nuclear proteinChain: A: PDB Molecule: kin17;PDBTitle: high resolution crystal structure of the human kin172 c-terminal domain containing a kow motif3 kin17.
19 c2xhcA_
93.0
18
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of thermotoga maritima n-utilization substance g2 (nusg)
20 c1m1gB_
92.3
25
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of aquifex aeolicus n-utilization2 substance g (nusg), space group p2(1)
21 c4a19F_
not modelled
91.0
26
PDB header: ribosomeChain: F: PDB Molecule: rpl14;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
22 c3izcN_
not modelled
90.7
23
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
23 c3iz5N_
not modelled
90.3
23
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
24 c3izcG_
not modelled
89.9
24
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein rpl6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
25 c2e6zA_
not modelled
87.6
25
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
26 c4a1dE_
not modelled
83.9
35
PDB header: ribosomeChain: E: PDB Molecule: rpl6;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
27 c4a18N_
not modelled
82.2
30
PDB header: ribosomeChain: N: PDB Molecule: rpl27;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
28 c2e70A_
not modelled
78.6
31
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the fifth kow motif of human2 transcription elongation factor spt5
29 c3iz5G_
not modelled
74.8
32
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein l6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
30 c2xzmW_
not modelled
56.5
16
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s4;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
31 c3kbgA_
not modelled
49.5
12
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s4e;PDBTitle: crystal structure of the 30s ribosomal protein s4e from2 thermoplasma acidophilum. northeast structural genomics3 consortium target tar28.
32 c3iz6D_
not modelled
44.8
29
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein s4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
33 d1vhka1
not modelled
43.3
18
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like34 c3izca_
not modelled
42.1
25
PDB header: ribosomeChain: A: PDB Molecule: 60s ribosomal protein rpl1 (l1p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
35 c4a1aP_
not modelled
35.8
32
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l21;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
36 c3iz5a_
not modelled
35.4
28
PDB header: ribosomeChain: A: PDB Molecule: 60s ribosomal protein l1 (l1p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
37 d1nxza1
not modelled
30.1
15
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like38 c2dxcG_
not modelled
28.5
22
PDB header: hydrolaseChain: G: PDB Molecule: thiocyanate hydrolase subunit alpha;PDBTitle: recombinant thiocyanate hydrolase, fully-matured form
39 d1o12a1
not modelled
24.8
29
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: N-acetylglucosamine-6-phosphate deacetylase, NagA40 c3k7aM_
not modelled
22.8
15
PDB header: transcriptionChain: M: PDB Molecule: transcription initiation factor iib;PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
41 d1ng2a2
not modelled
22.8
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain42 c2kmmA_
not modelled
20.7
28
PDB header: hydrolaseChain: A: PDB Molecule: guanosine-3',5'-bis(diphosphate) 3'-PDBTitle: solution nmr structure of the tgs domain of pg1808 from2 porphyromonas gingivalis. northeast structural genomics3 consortium target pgr122a (418-481)
43 c3qz9D_
not modelled
20.3
33
PDB header: lyaseChain: D: PDB Molecule: co-type nitrile hydratase beta subunit;PDBTitle: crystal structure of co-type nitrile hydratase beta-y215f from2 pseudomonas putida.
44 d1wlpb2
not modelled
20.1
30
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain45 c3izbD_
not modelled
19.3
23
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein rps4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
46 c3gqhB_
not modelled
19.3
19
PDB header: viral proteinChain: B: PDB Molecule: preneck appendage protein;PDBTitle: crystal structure of the bacteriophage phi29 gene product2 12 c-terminal fragment
47 d1pfta_
not modelled
18.9
14
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain48 d1tova_
not modelled
18.9
29
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain49 d1jb0e_
not modelled
18.7
23
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Photosystem I accessory protein E (PsaE)50 d1dl6a_
not modelled
18.3
3
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain51 d1psea_
not modelled
18.0
36
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Photosystem I accessory protein E (PsaE)52 c2k6dA_
not modelled
16.9
30
PDB header: sh3 domain/ubiquitinChain: A: PDB Molecule: sh3 domain-containing kinase-binding protein 1;PDBTitle: cin85 sh3-c domain in complex with ubiquitin
53 d1ugpb_
not modelled
16.7
30
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain54 d1p7ba1
not modelled
15.1
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytoplasmic domain of inward rectifier potassium channel55 d1qp3a_
not modelled
14.5
36
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Photosystem I accessory protein E (PsaE)56 c3dclC_
not modelled
14.4
40
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: tm1086;PDBTitle: crystal structure of tm1086
57 d1k4us_
not modelled
14.4
35
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain58 c2ed0A_
not modelled
14.2
17
PDB header: signaling proteinChain: A: PDB Molecule: abl interactor 2;PDBTitle: solution structure of the sh3 domain of abl interactor 22 (abelson interactor 2)
59 c2eyxA_
not modelled
13.6
30
PDB header: signaling proteinChain: A: PDB Molecule: v-crk sarcoma virus ct10 oncogene homologPDBTitle: c-terminal sh3 domain of ct10-regulated kinase
60 c2gb5B_
not modelled
13.5
8
PDB header: hydrolaseChain: B: PDB Molecule: nadh pyrophosphatase;PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
61 d1v29b_
not modelled
13.3
11
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain62 d2dk3a1
not modelled
12.8
38
Fold: SH3-like barrelSuperfamily: Mib/herc2 domain-likeFamily: Mib/herc2 domain63 c2l0aA_
not modelled
12.4
17
PDB header: signaling proteinChain: A: PDB Molecule: signal transducing adapter molecule 1;PDBTitle: solution nmr structure of signal transducing adapter molecule 1 stam-12 from homo sapiens, northeast structural genomics consortium target3 hr4479e
64 d2cona1
not modelled
12.4
25
Fold: Rubredoxin-likeSuperfamily: NOB1 zinc finger-likeFamily: NOB1 zinc finger-like65 d2c78a1
not modelled
12.2
24
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors66 c2drmB_
not modelled
11.8
39
PDB header: contractile proteinChain: B: PDB Molecule: acanthamoeba myosin ib;PDBTitle: acanthamoeba myosin i sh3 domain bound to acan125
67 c2dnuA_
not modelled
11.8
22
PDB header: structural genomics, structural proteinChain: A: PDB Molecule: sh3 multiple domains 1;PDBTitle: solution structure of rsgi ruh-061, a sh3 domain from human
68 c2xzm1_
not modelled
11.8
31
PDB header: ribosomeChain: 1: PDB Molecule: ribosomal protein s28e containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
69 d1xl4a1
not modelled
11.7
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytoplasmic domain of inward rectifier potassium channel70 d1ne3a_
not modelled
11.2
38
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like71 d1nlqa_
not modelled
11.1
9
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Nucleoplasmin-like core domainFamily: Nucleoplasmin-like core domain72 d1ppya_
not modelled
10.9
24
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC73 d1ny4a_
not modelled
10.8
46
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like74 d1gria2
not modelled
10.8
30
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain75 d1ka5a_
not modelled
10.7
28
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like76 c2csqA_
not modelled
10.7
14
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: rim binding protein 2;PDBTitle: solution structure of the second sh3 domain of human rim-2 binding protein 2
77 c1x2qA_
not modelled
10.6
22
PDB header: signaling proteinChain: A: PDB Molecule: signal transducing adapter molecule 2;PDBTitle: solution structure of the sh3 domain of the signal2 transducing adaptor molecule 2
78 d2hpra_
not modelled
10.5
22
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like79 c1wi7A_
not modelled
10.3
26
PDB header: protein bindingChain: A: PDB Molecule: sh3-domain kinase binding protein 1;PDBTitle: solution structure of the sh3 domain of sh3-domain kinase2 binding protein 1
80 c3t07D_
not modelled
10.3
19
PDB header: transferase/transferase inhibitorChain: D: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
81 c1u4fD_
not modelled
10.1
25
PDB header: allergenChain: D: PDB Molecule: inward rectifier potassium channel 2;PDBTitle: crystal structure of cytoplasmic domains of irk1 (kir2.1)2 channel
82 d2b1ya1
not modelled
10.0
26
Fold: Atu1913-likeSuperfamily: Atu1913-likeFamily: Atu1913-like83 c2bz8B_
not modelled
10.0
22
PDB header: sh3 domainChain: B: PDB Molecule: sh3-domain kinase binding protein 1;PDBTitle: n-terminal sh3 domain of cin85 bound to cbl-b peptide
84 d1n9pa_
not modelled
9.8
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytoplasmic domain of inward rectifier potassium channel85 d1mxaa2
not modelled
9.8
40
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase86 d1cm3a_
not modelled
9.7
17
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like87 c3ihsB_
not modelled
9.6
22
PDB header: transport proteinChain: B: PDB Molecule: phosphocarrier protein hpr;PDBTitle: crystal structure of a phosphocarrier protein hpr from2 bacillus anthracis str. ames
88 d2nzul1
not modelled
9.5
22
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like89 c3iz5U_
not modelled
9.5
31
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
90 c1wxbA_
not modelled
9.4
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: epidermal growth factor receptor pathwayPDBTitle: solution structure of the sh3 domain from human epidermal2 growth factor receptor pathway substrate 8-like protein
91 c2zkrq_
not modelled
9.1
36
PDB header: ribosomal protein/rnaChain: Q: PDB Molecule: rna expansion segment es31 part ii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
92 d1uhfa_
not modelled
8.9
22
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain93 c2bzxA_
not modelled
8.8
22
PDB header: sh3 domainChain: A: PDB Molecule: crk-like protein;PDBTitle: atomic model of crkl-sh3c monomer
94 d1ug1a_
not modelled
8.8
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain95 c2djqA_
not modelled
8.7
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 domain containing ring finger 2;PDBTitle: the solution structure of the first sh3 domain of mouse sh32 domain containing ring finger 2
96 d1ptfa_
not modelled
8.7
22
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like97 d1wlpb1
not modelled
8.6
13
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain98 c2wh7A_
not modelled
8.3
26
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronidase-phage associated;PDBTitle: the partial structure of a group a streptpcoccal phage-2 encoded tail fibre hyaluronate lyase hylp2
99 c2e4fA_
not modelled
8.2
18
PDB header: transport proteinChain: A: PDB Molecule: g protein-activated inward rectifier potassium channel 2;PDBTitle: crystal structure of the cytoplasmic domain of g-protein-gated inward2 rectifier potassium channel kir3.2