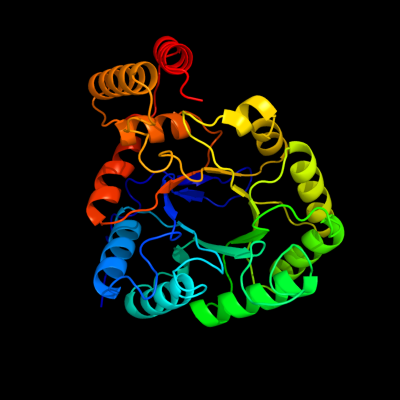
| 1 | c3n6qF_
|
|
 |
100.0 |
100 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:yghz aldo-keto reductase;
PDBTitle: crystal structure of yghz from e. coli
|
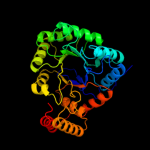
| 2 | c3erpA_
|
|
 |
100.0 |
61 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of idp01002, a putative oxidoreductase from and essential2 gene of salmonella typhimurium
|
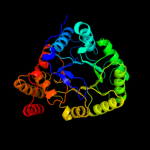
| 3 | d1lqaa_
|
|
 |
100.0 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
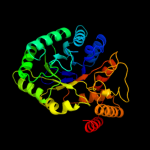
| 4 | d3eaua1
|
|
 |
100.0 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 5 | c3lutA_
|
|
 |
100.0 |
35 |
PDB header:membrane protein
Chain: A: PDB Molecule:voltage-gated potassium channel subunit beta-2;
PDBTitle: a structural model for the full-length shaker potassium channel kv1.2
|
| 6 | d1pyfa_
|
|
 |
100.0 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 7 | d1pz1a_
|
|
 |
100.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 8 | d1ur3m_
|
|
 |
100.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 9 | c3n2tA_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the glycerol dehydrogenase akr11b4 from gluconobacter2 oxydans
|
| 10 | d1gvea_
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 11 | c2bp1C_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:aflatoxin b1 aldehyde reductase member 2;
PDBTitle: structure of the aflatoxin aldehyde reductase in complex2 with nadph
|
| 12 | c1ynpA_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: aldo-keto reductase akr11c1 from bacillus halodurans (apo form)
|
| 13 | d1q5ma_
|
|
 |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 14 | d1j96a_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 15 | c3h7uA_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of the plant stress-response enzyme akr4c9
|
| 16 | d1afsa_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 17 | c3up8B_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative 2,5-diketo-d-gluconic acid reductase b;
PDBTitle: crystal structure of a putative 2,5-diketo-d-gluconic acid reductase b
|
| 18 | d1us0a_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 19 | d1qwka_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 20 | d1mi3a_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 21 | d1frba_ |
|
not modelled |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 22 | d1hqta_ |
|
not modelled |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 23 | c3f7jB_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:yvgn protein;
PDBTitle: b.subtilis yvgn
|
| 24 | d1s1pa_ |
|
not modelled |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 25 | c3h7rA_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of the plant stress-response enzyme akr4c8
|
| 26 | c3buvB_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxo-5-beta-steroid 4-dehydrogenase;
PDBTitle: crystal structure of human delta(4)-3-ketosteroid 5-beta-reductase in2 complex with nadp and hepes. resolution: 1.35 a.
|
| 27 | d1ah4a_ |
|
not modelled |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 28 | c1zgdB_ |
|
not modelled |
100.0 |
23 |
PDB header:plant protein
Chain: B: PDB Molecule:chalcone reductase;
PDBTitle: chalcone reductase complexed with nadp+ at 1.7 angstrom2 resolution
|
| 29 | c2wztA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of a mycobacterium aldo-keto reductase in2 its apo and liganded form
|
| 30 | d1c9wa_ |
|
not modelled |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 31 | d1vp5a_ |
|
not modelled |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 32 | d2alra_ |
|
not modelled |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 33 | d1hw6a_ |
|
not modelled |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 34 | c3o0kB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aldo/keto reductase;
PDBTitle: crystal structure of aldo/keto reductase from brucella melitensis
|
| 35 | d1mzra_ |
|
not modelled |
100.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 36 | c2bgsA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldose reductase;
PDBTitle: holo aldose reductase from barley
|
| 37 | c3b3dA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative morphine dehydrogenase;
PDBTitle: b.subtilis ytbe
|
| 38 | c1vbjB_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin f synthase;
PDBTitle: the crystal structure of prostaglandin f synthase from2 trypanosoma brucei
|
| 39 | c3krbB_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aldose reductase;
PDBTitle: structure of aldose reductase from giardia lamblia at 1.75a resolution
|
| 40 | c3ln3A_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodiol dehydrogenase;
PDBTitle: crystal structure of putative reductase (np_038806.2) from2 mus musculus at 1.18 a resolution
|
| 41 | d3bofa2 |
|
not modelled |
73.3 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 42 | c3dx5A_ |
|
not modelled |
60.8 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 43 | c3bdkB_ |
|
not modelled |
54.4 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
| 44 | c3bolB_ |
|
not modelled |
52.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 45 | c3sb1B_ |
|
not modelled |
38.0 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hydrogenase expression protein;
PDBTitle: hydrogenase expression protein huph from thiobacillus denitrificans2 atcc 25259
|
| 46 | c3oqbF_ |
|
not modelled |
37.0 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from bradyrhizobium2 japonicum usda 110
|
| 47 | c3hf3A_ |
|
not modelled |
35.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 48 | d1szna2 |
|
not modelled |
33.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 49 | c3a5vA_ |
|
not modelled |
33.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of alpha-galactosidase i from mortierella vinacea
|
| 50 | c3g8rA_ |
|
not modelled |
32.5 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable spore coat polysaccharide biosynthesis protein e;
PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
|
| 51 | c2ph5A_ |
|
not modelled |
31.6 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
| 52 | c3ou8B_ |
|
not modelled |
31.1 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenosine deaminase;
PDBTitle: the crystal structure of adenosine deaminase from pseudomonas2 aeruginosa
|
| 53 | d1ajza_ |
|
not modelled |
30.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 54 | c1uasA_ |
|
not modelled |
29.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of rice alpha-galactosidase
|
| 55 | c3rysA_ |
|
not modelled |
29.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosine deaminase 1;
PDBTitle: the crystal structure of adenine deaminase (aaur1117) from2 arthrobacter aurescens
|
| 56 | d1jpma1 |
|
not modelled |
26.2 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 57 | d1gqna_ |
|
not modelled |
25.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 58 | d1h8ba_ |
|
not modelled |
23.1 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:EF-hand modules in multidomain proteins |
| 59 | d2nlya1 |
|
not modelled |
22.2 |
15 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:Divergent polysaccharide deacetylase |
| 60 | d1vp8a_ |
|
not modelled |
19.5 |
14 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 61 | d1bxba_ |
|
not modelled |
19.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 62 | c1m0tB_ |
|
not modelled |
19.3 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:glutathione synthetase;
PDBTitle: yeast glutathione synthase
|
| 63 | d1tz9a_ |
|
not modelled |
19.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:UxuA-like |
| 64 | d1qt1a_ |
|
not modelled |
18.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 65 | d1zy9a2 |
|
not modelled |
18.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:YicI catalytic domain-like |
| 66 | c2kzhA_ |
|
not modelled |
17.1 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:tryptophan biosynthesis protein trpcf;
PDBTitle: three-dimensional structure of a truncated phosphoribosylanthranilate2 isomerase (residues 255-384) from escherichia coli
|
| 67 | c3d0cB_ |
|
not modelled |
17.1 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
|
| 68 | d1kkoa1 |
|
not modelled |
17.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 69 | d2hv2a1 |
|
not modelled |
17.0 |
21 |
Fold:SCP-like
Superfamily:SCP-like
Family:EF1021 C-terminal domain-like |
| 70 | c3ksmA_ |
|
not modelled |
16.8 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
| 71 | c2yr1B_ |
|
not modelled |
16.3 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
|
| 72 | d2i00a1 |
|
not modelled |
16.2 |
14 |
Fold:SCP-like
Superfamily:SCP-like
Family:EF1021 C-terminal domain-like |
| 73 | c2nvaH_ |
|
not modelled |
16.0 |
15 |
PDB header:lyase
Chain: H: PDB Molecule:arginine decarboxylase, a207r protein;
PDBTitle: the x-ray crystal structure of the paramecium bursaria2 chlorella virus arginine decarboxylase bound to agmatine
|
| 74 | c2gjxE_ |
|
not modelled |
16.0 |
17 |
PDB header:hydrolase
Chain: E: PDB Molecule:beta-hexosaminidase alpha chain;
PDBTitle: crystallographic structure of human beta-hexosaminidase a
|
| 75 | c3dnfB_ |
|
not modelled |
15.7 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
|
| 76 | d1uwka_ |
|
not modelled |
15.5 |
16 |
Fold:Urocanase
Superfamily:Urocanase
Family:Urocanase |
| 77 | c2ekcA_ |
|
not modelled |
14.5 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 78 | c3sdoB_ |
|
not modelled |
14.4 |
4 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
| 79 | d2gjxa1 |
|
not modelled |
14.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 80 | d1vyra_ |
|
not modelled |
14.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 81 | d2ijqa1 |
|
not modelled |
14.1 |
10 |
Fold:Hyaluronidase domain-like
Superfamily:TTHA0068-like
Family:TTHA0068-like |
| 82 | c2wjeA_ |
|
not modelled |
13.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase cpsb;
PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
|
| 83 | c2vwtA_ |
|
not modelled |
13.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|
| 84 | c2ksnA_ |
|
not modelled |
13.5 |
31 |
PDB header:signaling protein
Chain: A: PDB Molecule:ubiquitin domain-containing protein 2;
PDBTitle: solution structure of the n-terminal domain of dc-ubp/ubtd2
|
| 85 | d1ub3a_ |
|
not modelled |
13.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 86 | c2fknC_ |
|
not modelled |
12.6 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:urocanate hydratase;
PDBTitle: crystal structure of urocanase from bacillus subtilis
|
| 87 | c1t0oA_ |
|
not modelled |
11.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: the structure of alpha-galactosidase from trichoderma reesei complexed2 with beta-d-galactose
|
| 88 | c2v5jB_ |
|
not modelled |
11.8 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;
PDBTitle: apo class ii aldolase hpch
|
| 89 | c1vliA_ |
|
not modelled |
11.5 |
20 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
| 90 | c1xeaD_ |
|
not modelled |
11.3 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
| 91 | c3pueA_ |
|
not modelled |
11.1 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 92 | c3lmyA_ |
|
not modelled |
10.8 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase subunit beta;
PDBTitle: the crystal structure of beta-hexosaminidase b in complex with2 pyrimethamine
|
| 93 | d2zd1b1 |
|
not modelled |
10.8 |
21 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Reverse transcriptase |
| 94 | d2ajta2 |
|
not modelled |
10.7 |
9 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:AraA N-terminal and middle domain-like |
| 95 | d1x87a_ |
|
not modelled |
10.2 |
16 |
Fold:Urocanase
Superfamily:Urocanase
Family:Urocanase |
| 96 | c3l2iB_ |
|
not modelled |
10.2 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
| 97 | c2jwlB_ |
|
not modelled |
10.1 |
16 |
PDB header:membrane protein
Chain: B: PDB Molecule:protein tolr;
PDBTitle: solution structure of periplasmic domain of tolr from h.2 influenzae with saxs data
|
| 98 | c3gh7A_ |
|
not modelled |
10.1 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of beta-hexosaminidase from paenibacillus2 sp. ts12 in complex with galnac
|
| 99 | d1nowa1 |
|
not modelled |
10.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |