| 1 | c3nreB_ |
|
 |
100.0 |
20 |
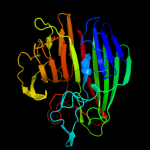
PDB header:isomerase
Chain: B: PDB Molecule:aldose 1-epimerase;
PDBTitle: crystal structure of a putative aldose 1-epimerase (b2544) from2 escherichia coli k12 at 1.59 a resolution
|
| 2 | d1z45a1 |
|
 |
100.0 |
23 |
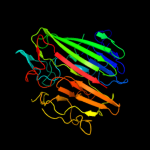
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Aldose 1-epimerase (mutarotase) |
| 3 | d1so0a_ |
|
 |
100.0 |
23 |
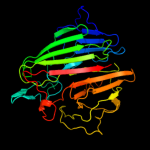
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Aldose 1-epimerase (mutarotase) |
| 4 | c3imhB_ |
|
 |
100.0 |
19 |
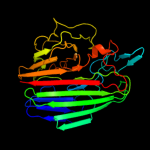
PDB header:isomerase
Chain: B: PDB Molecule:galactose-1-epimerase;
PDBTitle: crystal structure of galactose 1-epimerase from lactobacillus2 acidophilus ncfm
|
| 5 | c1ygaA_ |
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:hypothetical 37.9 kda protein in bio3-hxt17
PDBTitle: crystal structure of saccharomyces cerevisiae yn9a protein,2 new york structural genomics consortium
|
| 6 | c1z45A_ |
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:gal10 bifunctional protein;
PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces3 cerevisiae complexed with nad, udp-glucose, and galactose
|
| 7 | d1lura_ |
|
 |
100.0 |
22 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Aldose 1-epimerase (mutarotase) |
| 8 | d1nsza_ |
|
 |
100.0 |
20 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Aldose 1-epimerase (mutarotase) |
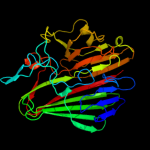
| 9 | c3os7B_ |
|
 |
100.0 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:galactose mutarotase-like protein;
PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
|
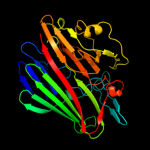
| 10 | c3os7D_ |
|
 |
100.0 |
19 |
PDB header:isomerase
Chain: D: PDB Molecule:galactose mutarotase-like protein;
PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
|
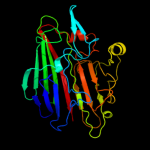
| 11 | c3dcdA_ |
|
 |
100.0 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:galactose mutarotase related enzyme;
PDBTitle: x-ray structure of the galactose mutarotase related enzyme q5fkd7 from2 lactobacillus acidophilus at the resolution 1.9a. northeast3 structural genomics consortium target lar33.
|
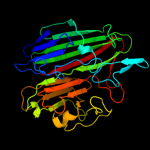
| 12 | c3q1nA_ |
|
 |
100.0 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:galactose mutarotase related enzyme;
PDBTitle: crystal structure of a galactose mutarotase-like protein (lsei_2598)2 from lactobacillus casei atcc 334 at 1.61 a resolution
|
| 13 | c3mwxA_ |
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:aldose 1-epimerase;
PDBTitle: crystal structure of a putative galactose mutarotase (bsu18360) from2 bacillus subtilis at 1.45 a resolution
|
| 14 | c3k25B_ |
|
 |
100.0 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:slr1438 protein;
PDBTitle: crystal structure of slr1438 protein from synechocystis sp. pcc 6803,2 northeast structural genomics consortium target sgr112
|
| 15 | c2cisA_ |
|
 |
100.0 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:hexose-6-phosphate mutarotase;
PDBTitle: structure-based functional annotation: yeast ymr099c codes2 for a d-hexose-6-phosphate mutarotase. complex with3 tagatose-6-phosphate
|
| 16 | c2htbB_ |
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:putative enzyme related to aldose 1-epimerase;
PDBTitle: crystal structure of a putative mutarotase (yead) from2 salmonella typhimurium in monoclinic form
|
| 17 | d1jova_ |
|
 |
100.0 |
18 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Hypothetical protein HI1317 |
| 18 | c3ty1B_ |
|
 |
100.0 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:hypothetical aldose 1-epimerase;
PDBTitle: crystal structure of a hypothetical aldose 1-epimerase (kpn_04629)2 from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 1.90 a3 resolution
|
| 19 | c3blcB_ |
|
 |
98.1 |
12 |
PDB header:chaperone,protein transport
Chain: B: PDB Molecule:inner membrane protein oxaa;
PDBTitle: crystal structure of the periplasmic domain of the escherichia coli2 yidc
|
| 20 | c3bs6B_ |
|
 |
98.1 |
10 |
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:inner membrane protein oxaa;
PDBTitle: 1.8 angstrom crystal structure of the periplasmic domain of2 the membrane insertase yidc
|
| 21 | d1jz8a4 |
|
not modelled |
93.2 |
12 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:beta-Galactosidase, domain 5 |
| 22 | c2yfnA_ |
|
not modelled |
89.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase-sucrose kinase agask;
PDBTitle: galactosidase domain of alpha-galactosidase-sucrose kinase,2 agask
|
| 23 | c2xn1B_ |
|
not modelled |
89.0 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-galactosidase;
PDBTitle: structure of alpha-galactosidase from lactobacillus acidophilus ncfm2 with tris
|
| 24 | c3rgbA_ |
|
not modelled |
86.3 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methane monooxygenase subunit b2;
PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
|
| 25 | c1yewI_ |
|
not modelled |
86.3 |
14 |
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
|
| 26 | c3mi6A_ |
|
not modelled |
83.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of the alpha-galactosidase from lactobacillus2 brevis, northeast structural genomics consortium target lbr11.
|
| 27 | c3rfrI_ |
|
not modelled |
82.2 |
17 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:pmob;
PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
|
| 28 | c3bgaB_ |
|
not modelled |
75.3 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of beta-galactosidase from bacteroides2 thetaiotaomicron vpi-5482
|
| 29 | c1jz6C_ |
|
not modelled |
74.5 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-galactosidase;
PDBTitle: e. coli (lacz) beta-galactosidase in complex with galacto-2 tetrazole
|
| 30 | c3mv14_ |
|
not modelled |
71.1 |
14 |
PDB header:hydrolase
Chain: 4: PDB Molecule:beta-galactosidase;
PDBTitle: e.coli (lacz) beta-galactosidase (r599a) in complex with guanidinium
|
| 31 | c1so9A_ |
|
not modelled |
65.5 |
10 |
PDB header:metal transport
Chain: A: PDB Molecule:cytochrome c oxidase assembly protein ctag;
PDBTitle: solution structure of apocox11, 30 structures
|
| 32 | d1so9a_ |
|
not modelled |
65.5 |
10 |
Fold:Ctag/Cox11
Superfamily:Ctag/Cox11
Family:Ctag/Cox11 |
| 33 | d1ejxb_ |
|
not modelled |
63.0 |
17 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 34 | d4ubpb_ |
|
not modelled |
62.3 |
26 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 35 | d1e9ya1 |
|
not modelled |
52.4 |
22 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 36 | c3qgaD_ |
|
not modelled |
50.7 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:fusion of urease beta and gamma subunits;
PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
|
| 37 | d1ji1a2 |
|
not modelled |
49.2 |
8 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 38 | c3qbtH_ |
|
not modelled |
47.2 |
26 |
PDB header:protein transport/hydrolase
Chain: H: PDB Molecule:inositol polyphosphate 5-phosphatase ocrl-1;
PDBTitle: crystal structure of ocrl1 540-678 in complex with rab8a:gppnhp
|
| 39 | c1e9zA_ |
|
not modelled |
44.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:urease subunit alpha;
PDBTitle: crystal structure of helicobacter pylori urease
|
| 40 | c3obaA_ |
|
not modelled |
39.7 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: structure of the beta-galactosidase from kluyveromyces lactis
|
| 41 | d1nkga3 |
|
not modelled |
38.0 |
11 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Rhamnogalacturonase B, RhgB, N-terminal domain |
| 42 | c1nkgA_ |
|
not modelled |
32.3 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:rhamnogalacturonase b;
PDBTitle: rhamnogalacturonan lyase from aspergillus aculeatus
|
| 43 | d1wzla2 |
|
not modelled |
25.0 |
17 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 44 | c3isyA_ |
|
not modelled |
24.4 |
6 |
PDB header:protein binding
Chain: A: PDB Molecule:intracellular proteinase inhibitor;
PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
|
| 45 | c2r39A_ |
|
not modelled |
19.8 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:fixg-related protein;
PDBTitle: crystal structure of fixg-related protein from vibrio parahaemolyticus
|
| 46 | c2x3bB_ |
|
not modelled |
19.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:toxic extracellular endopeptidase;
PDBTitle: asap1 inactive mutant e294a, an extracellular toxic zinc2 metalloendopeptidase
|
| 47 | c3ac0B_ |
|
not modelled |
16.7 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase i;
PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
|
| 48 | c2cqtA_ |
|
not modelled |
16.2 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:cellobiose phosphorylase;
PDBTitle: crystal structure of cellvibrio gilvus cellobiose phosphorylase2 crystallized from sodium/potassium phosphate
|
| 49 | d2je8a2 |
|
not modelled |
15.1 |
12 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 50 | c2zf9D_ |
|
not modelled |
12.6 |
15 |
PDB header:structural protein
Chain: D: PDB Molecule:scae cell-surface anchored scaffoldin protein;
PDBTitle: crystal structure of a type iii cohesin module from the cellulosomal2 scae cell-surface anchoring scaffoldin of ruminococcus flavefaciens
|
| 51 | c2kl8A_ |
|
not modelled |
11.5 |
31 |
PDB header:de novo protein
Chain: A: PDB Molecule:or15;
PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
|
| 52 | d1cc3a_ |
|
not modelled |
10.7 |
20 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 53 | d1g94a1 |
|
not modelled |
10.4 |
21 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 54 | d1cx1a_ |
|
not modelled |
9.8 |
18 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:CBM4/9 |
| 55 | d1nbca_ |
|
not modelled |
9.0 |
15 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
| 56 | c3cfuA_ |
|
not modelled |
8.7 |
33 |
PDB header:lipoprotein
Chain: A: PDB Molecule:uncharacterized lipoprotein yjha;
PDBTitle: crystal structure of the yjha protein from bacillus2 subtilis. northeast structural genomics consortium target3 sr562
|
| 57 | c1v7wA_ |
|
not modelled |
8.6 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:chitobiose phosphorylase;
PDBTitle: crystal structure of vibrio proteolyticus chitobiose phosphorylase in2 complex with glcnac
|
| 58 | d1gu3a_ |
|
not modelled |
8.1 |
18 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:CBM4/9 |
| 59 | d1v7wa2 |
|
not modelled |
8.1 |
12 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Glycosyltransferase family 36 N-terminal domain |
| 60 | d1ktba1 |
|
not modelled |
8.0 |
14 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 61 | d1g43a_ |
|
not modelled |
7.9 |
5 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
| 62 | c3zqwA_ |
|
not modelled |
7.7 |
15 |
PDB header:carbohydrate-binding protein
Chain: A: PDB Molecule:cellulosomal scaffoldin;
PDBTitle: structure of cbm3b of major scaffoldin subunit scaa from2 acetivibrio cellulolyticus
|
| 63 | c2l8aA_ |
|
not modelled |
7.6 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase;
PDBTitle: structure of a novel cbm3 lacking the calcium-binding site
|
| 64 | d1dkza2 |
|
not modelled |
7.4 |
23 |
Fold:Heat shock protein 70kD (HSP70), peptide-binding domain
Superfamily:Heat shock protein 70kD (HSP70), peptide-binding domain
Family:Heat shock protein 70kD (HSP70), peptide-binding domain |
| 65 | d2ccwa1 |
|
not modelled |
7.3 |
17 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 66 | c3dqgC_ |
|
not modelled |
7.0 |
13 |
PDB header:chaperone
Chain: C: PDB Molecule:heat shock 70 kda protein f;
PDBTitle: peptide-binding domain of heat shock 70 kda protein f, mitochondrial2 precursor, from caenorhabditis elegans.
|
| 67 | d1u00a2 |
|
not modelled |
6.7 |
20 |
Fold:Heat shock protein 70kD (HSP70), peptide-binding domain
Superfamily:Heat shock protein 70kD (HSP70), peptide-binding domain
Family:Heat shock protein 70kD (HSP70), peptide-binding domain |
| 68 | c3qfgA_ |
|
not modelled |
6.6 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: structure of a putative lipoprotein from staphylococcus aureus subsp.2 aureus nctc 8325
|
| 69 | d1cuoa_ |
|
not modelled |
6.4 |
17 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 70 | d3dhpa1 |
|
not modelled |
6.4 |
19 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 71 | d1mspa_ |
|
not modelled |
6.4 |
22 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:PapD-like
Family:MSP-like |
| 72 | c2wo4A_ |
|
not modelled |
6.3 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycoside hydrolase, family 9;
PDBTitle: 3b' carbohydrate-binding module from the cel9v glycoside2 hydrolase from clostridium thermocellum, in-house data
|
| 73 | d2vzsa2 |
|
not modelled |
6.2 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 74 | d3es6b1 |
|
not modelled |
6.0 |
8 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:SVA-like |
| 75 | d2j5wa3 |
|
not modelled |
5.9 |
0 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 76 | d1hfua2 |
|
not modelled |
5.8 |
13 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 77 | d2f2ha2 |
|
not modelled |
5.8 |
8 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:YicI N-terminal domain-like |
| 78 | d1azca_ |
|
not modelled |
5.6 |
17 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 79 | c3tdqB_ |
|
not modelled |
5.6 |
19 |
PDB header:cell adhesion
Chain: B: PDB Molecule:pily2 protein;
PDBTitle: crystal structure of a fimbrial biogenesis protein pily22 (pily2_pa4555) from pseudomonas aeruginosa pao1 at 2.10 a resolution
|
| 80 | d1jaea1 |
|
not modelled |
5.2 |
16 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 81 | c2aenH_ |
|
not modelled |
5.2 |
12 |
PDB header:viral protein
Chain: H: PDB Molecule:outer capsid protein vp4, vp8* core;
PDBTitle: crystal structure of the rotavirus strain ds-1 vp8* core
|

