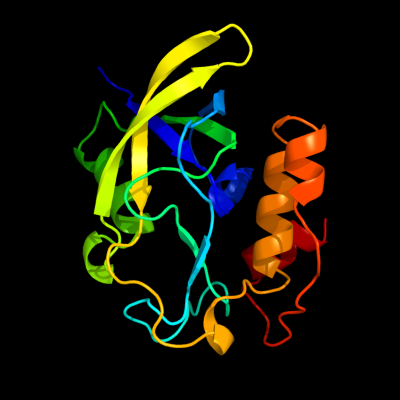
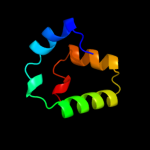
1 c2rbhA_
99.9
16
PDB header: transferaseChain: A: PDB Molecule: gamma-glutamyl cyclotransferase;PDBTitle: gamma-glutamyl cyclotransferase
2 c2qikA_
99.7
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0131 protein ykqa;PDBTitle: crystal structure of ykqa from bacillus subtilis. northeast2 structural genomics target sr631
3 c2g0qA_
98.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: at5g39720.1 protein;PDBTitle: solution structure of at5g39720.1 from arabidopsis thaliana
4 c2jqvA_
98.6
16
PDB header: structural genomicsChain: A: PDB Molecule: aig2 protein-like;PDBTitle: solution structure at3g28950.1 from arabidopsis thaliana
5 d1xhsa_
98.5
19
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like6 c3jubA_
98.2
20
PDB header: transferaseChain: A: PDB Molecule: aig2-like domain-containing protein 1;PDBTitle: human gamma-glutamylamine cyclotransferase
7 d1vkba_
98.1
23
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like8 d1v30a_
97.3
19
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like9 c2x8qA_
55.1
10
PDB header: virusChain: A: PDB Molecule: capsid protein p27;PDBTitle: cryo-em 3d model of the icosahedral particle2 composed of rous sarcoma virus capsid protein pentamers
10 d1em9a_
54.9
10
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain11 c2kgfA_
47.3
16
PDB header: viral proteinChain: A: PDB Molecule: capsid protein p27;PDBTitle: n-terminal domain of capsid protein from the mason-pfizer2 monkey virus
12 c2c2zB_
44.8
21
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: caspase-8 p10 subunit;PDBTitle: crystal structure of caspase-8 in complex with aza-peptide michael2 acceptor inhibitor
13 c3edyA_
36.5
24
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure of the precursor form of human tripeptidyl-peptidase2 1
14 d1p7na_
35.6
11
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain15 c3ee6A_
35.4
24
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure analysis of tripeptidyl peptidase -i
16 c3m66A_
34.5
7
PDB header: transcriptionChain: A: PDB Molecule: mterf domain-containing protein 1, mitochondrial;PDBTitle: crystal structure of human mitochondrial transcription termination2 factor 3
17 d1bdsa_
33.9
35
Fold: Defensin-likeSuperfamily: Defensin-likeFamily: Defensin18 d1gk8i_
28.4
21
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit19 d1d1da2
26.5
10
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain20 d2cvea2
24.4
24
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: YigZ C-terminal domain-like21 d2plga1
not modelled
22.5
44
Fold: Secretion chaperone-likeSuperfamily: Type III secretory system chaperone-likeFamily: Tll0839-like22 d1mpga1
not modelled
22.2
21
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: DNA repair glycosylase, 2 C-terminal domains23 d1fp2a1
not modelled
22.1
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain24 d1wdds_
not modelled
21.7
21
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit25 c2v4xA_
not modelled
21.5
8
PDB header: viral proteinChain: A: PDB Molecule: capsid protein p27;PDBTitle: crystal structure of jaagsiekte sheep retrovirus capsid n-2 terminal domain
26 d2v6ai1
not modelled
20.6
21
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit27 c2dwuA_
not modelled
20.1
16
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
28 d1no5a_
not modelled
20.1
25
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase29 d8ruci_
not modelled
19.1
21
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit30 c2jfoB_
not modelled
19.0
16
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
31 d1uzdc1
not modelled
18.7
21
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit32 c1mpgB_
not modelled
18.4
21
PDB header: hydrolaseChain: B: PDB Molecule: 3-methyladenine dna glycosylase ii;PDBTitle: 3-methyladenine dna glycosylase ii from escherichia coli
33 d1ej7s_
not modelled
18.2
21
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit34 c2qljB_
not modelled
17.6
23
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: caspase-7;PDBTitle: crystal structure of caspase-7 with inhibitor ac-wehd-cho
35 c3c7bA_
not modelled
17.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunit alpha;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
36 d1t1ea2
not modelled
16.3
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors37 d1mm0a_
not modelled
15.9
56
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Insect defensins38 c1b74A_
not modelled
15.6
24
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: glutamate racemase from aquifex pyrophilus
39 c3lehA_
not modelled
15.4
32
PDB header: transferaseChain: A: PDB Molecule: putative hydroxymethylglutaryl-coa synthase;PDBTitle: the crystal structure of smu.943c from streptococcus mutans ua159
40 d1p17b_
not modelled
15.0
33
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)41 c3hfrA_
not modelled
14.9
14
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
42 c1kmcB_
not modelled
14.6
23
PDB header: apoptosis/hydrolaseChain: B: PDB Molecule: caspase-7;PDBTitle: crystal structure of the caspase-7 / xiap-bir2 complex
43 c3degC_
not modelled
14.5
16
PDB header: ribosomeChain: C: PDB Molecule: gtp-binding protein lepa;PDBTitle: complex of elongating escherichia coli 70s ribosome and ef4(lepa)-2 gmppnp
44 d1ucva_
not modelled
14.2
13
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain45 c3nwuB_
not modelled
14.1
16
PDB header: hydrolaseChain: B: PDB Molecule: serine protease htra1;PDBTitle: substrate induced remodeling of the active site regulates htra12 activity
46 d1pzma_
not modelled
14.1
33
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)47 c2p2cD_
not modelled
14.0
24
PDB header: hydrolaseChain: D: PDB Molecule: caspase-2;PDBTitle: inhibition of caspase-2 by a designed ankyrin repeat2 protein (darpin)
48 d2ibna1
not modelled
13.9
29
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: MioX-like49 c2ohoA_
not modelled
13.8
27
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: structural basis for glutamate racemase inhibitor
50 c3kopB_
not modelled
13.3
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
51 d1tc1a_
not modelled
13.3
33
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)52 c3d0wD_
not modelled
13.3
24
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: yflh protein;PDBTitle: crystal structure of yflh protein from bacillus subtilis.2 northeast structural genomics consortium target sr326
53 d1riqa1
not modelled
12.8
21
Fold: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)Superfamily: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)Family: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)54 c2ywtA_
not modelled
12.2
33
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: crystal structure of hypoxanthine-guanine2 phosphoribosyltransferase with gmp from thermus3 thermophilus hb8
55 c3my3A_
not modelled
12.1
4
PDB header: transcriptionChain: A: PDB Molecule: mterf domain-containing protein 1, mitochondrial;PDBTitle: crystal structure of human mitochondrial transcription termination2 factor 3
56 d1y88a1
not modelled
11.8
14
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: Hypothetical protein AF1548, C-terminal domain57 d2qf3a1
not modelled
11.6
13
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Prokaryotic proteases58 c2v4wB_
not modelled
11.5
32
PDB header: transferaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa synthase,PDBTitle: crystal structure of human mitochondrial 3-hydroxy-3-2 methylglutaryl-coenzyme a synthase 2 (hmgcs2)
59 c3n6sA_
not modelled
11.4
14
PDB header: transcription, replication/dnaChain: A: PDB Molecule: transcription termination factor, mitochondrial;PDBTitle: crystal structure of human mitochondrial mterf in complex with a 15-2 mer dna encompassing the trnaleu(uur) binding sequence
60 c3kb8A_
not modelled
10.9
33
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: 2.09 angstrom resolution structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-1) from bacillus anthracis str. 'ames3 ancestor' in complex with gmp
61 c1ru7B_
not modelled
10.8
28
PDB header: viral proteinChain: B: PDB Molecule: hemagglutinin;PDBTitle: 1934 human h1 hemagglutinin
62 c2c1eB_
not modelled
10.8
17
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: caspase-3 subunit p12;PDBTitle: crystal structures of caspase-3 in complex with aza-peptide michael2 acceptor inhibitors.
63 d1g9sa_
not modelled
10.8
33
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)64 c2dl0A_
not modelled
10.7
16
PDB header: signaling proteinChain: A: PDB Molecule: sam and sh3 domain-containing protein 1;PDBTitle: solution structure of the sam-domain of the sam and sh32 domain containing protein 1
65 c1x9eB_
not modelled
10.6
27
PDB header: lyaseChain: B: PDB Molecule: hmg-coa synthase;PDBTitle: crystal structure of hmg-coa synthase from enterococcus2 faecalis
66 c2jfzB_
not modelled
10.5
16
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
67 c3bt6B_
not modelled
10.4
25
PDB header: viral proteinChain: B: PDB Molecule: influenza b hemagglutinin (ha);PDBTitle: crystal structure of influenza b virus hemagglutinin
68 c3o7mD_
not modelled
10.4
50
PDB header: transferaseChain: D: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: 1.98 angstrom resolution crystal structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-2) from bacillus anthracis str. 'ames3 ancestor'
69 c2h51B_
not modelled
10.3
6
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: caspase-1;PDBTitle: crystal structure of human caspase-1 (glu390->asp and arg286->lys) in2 complex with 3-[2-(2-benzyloxycarbonylamino-3-methyl-butyrylamino)-3 propionylamino]-4-oxo-pentanoic acid (z-vad-fmk)
70 c2gzmB_
not modelled
10.0
16
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of the glutamate racemase from bacillus2 anthracis
71 c3outC_
not modelled
9.9
19
PDB header: isomeraseChain: C: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
72 d1fp1d1
not modelled
9.7
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain73 d1pgw21
not modelled
9.6
40
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP74 d1pgl21
not modelled
9.5
40
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP75 d1ny721
not modelled
9.5
40
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP76 c2jbhA_
not modelled
9.3
22
PDB header: transferaseChain: A: PDB Molecule: phosphoribosyltransferase domain-containing protein 1;PDBTitle: human phosphoribosyl transferase domain containing 1
77 d1hgxa_
not modelled
9.3
50
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)78 d1ccwa_
not modelled
9.2
10
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain79 c2wrhI_
not modelled
9.2
28
PDB header: viral proteinChain: I: PDB Molecule: hemagglutinin ha2 chain;PDBTitle: structure of h1 duck albert hemagglutinin with human2 receptor
80 c1hgeD_
not modelled
9.1
22
PDB header: viral proteinChain: D: PDB Molecule: hemagglutinin, (g135r), ha1 chain;PDBTitle: binding of influenza virus hemagglutinin to analogs of its cell-2 surface receptor, sialic acid: analysis by proton nuclear magnetic3 resonance spectroscopy and x-ray crystallography
81 c3s5oA_
not modelled
9.1
12
PDB header: lyaseChain: A: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase, mitochondrial;PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
82 c3m5gD_
not modelled
9.1
28
PDB header: viral proteinChain: D: PDB Molecule: hemagglutinin;PDBTitle: crystal structure of a h7 influenza virus hemagglutinin
83 c1jsdB_
not modelled
9.1
31
PDB header: viral proteinChain: B: PDB Molecule: haemagglutinin (ha2 chain);PDBTitle: crystal structure of swine h9 haemagglutinin
84 c3sirD_
not modelled
9.1
23
PDB header: hydrolaseChain: D: PDB Molecule: caspase;PDBTitle: crystal structure of drice
85 d1zs4a1
not modelled
9.1
26
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Bacteriophage CII protein86 c2ja9A_
not modelled
8.9
8
PDB header: rna-binding proteinChain: A: PDB Molecule: exosome complex exonuclease rrp40;PDBTitle: structure of the n-terminal deletion of yeast exosome2 component rrp40
87 c2p8uB_
not modelled
8.8
45
PDB header: transferaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa synthase, cytoplasmic;PDBTitle: crystal structure of human 3-hydroxy-3-methylglutaryl coa synthase i
88 d1f1ja_
not modelled
8.6
23
Fold: Caspase-likeSuperfamily: Caspase-likeFamily: Caspase catalytic domain89 c2d9xA_
not modelled
8.5
17
PDB header: lipid transportChain: A: PDB Molecule: oxysterol binding protein-related protein 11;PDBTitle: solution structure of the ph domain of oxysterol binding2 protein-related protein 11 from human
90 d1nu0a_
not modelled
8.4
35
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX91 d1o5ka_
not modelled
8.3
28
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase92 c2rq5A_
not modelled
8.3
21
PDB header: transcriptionChain: A: PDB Molecule: protein jumonji;PDBTitle: solution structure of the at-rich interaction domain (arid)2 of jumonji/jarid2
93 c3nziA_
not modelled
8.3
21
PDB header: hydrolase/hydrolase substrateChain: A: PDB Molecule: serine protease htra1;PDBTitle: substrate induced remodeling of the active site regulates htra12 activity
94 c3h9xB_
not modelled
8.3
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein pspto_3016;PDBTitle: crystal structure of the pspto_3016 protein from2 pseudomonas syringae, northeast structural genomics3 consortium target psr293
95 c2funB_
not modelled
8.2
24
PDB header: apoptosis/hydrolaseChain: B: PDB Molecule: caspase-8;PDBTitle: alternative p35-caspase-8 complex
96 c1htmB_
not modelled
8.2
22
PDB header: viral proteinChain: B: PDB Molecule: hemagglutinin ha2 chain;PDBTitle: structure of influenza haemagglutinin at the ph of membrane2 fusion
97 d2ia7a1
not modelled
7.9
20
Fold: gpW/gp25-likeSuperfamily: gpW/gp25-likeFamily: gpW/gp25-like98 c3n7qA_
not modelled
7.9
14
PDB header: transcription, replication/dnaChain: A: PDB Molecule: transcription termination factor, mitochondrial;PDBTitle: crystal structure of human mitochondrial mterf fragment (aa 99-399) in2 complex with a 12-mer dna encompassing the trnaleu(uur) binding3 sequence
99 c1yfzA_
not modelled
7.9
33
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis